Tutorial - FHIR ImagingStudy with EHR data (Duke University data)
Tutorial code on GitHub hapi-py-fhir-tutorials -- dukeunidata.py.
Currently, it is a private repository, will open source later...
Background
Scenario
Last time we mentioned the volunteer Linman took the breast MRIs at the Akron Hospital.The researcher at BioResearch Institute stored the collected MRI data in SPARC SDS datasets. And also he record some interest data in the below form that he would use to do further research. And our aim is to store these EHR data into FHIR resource.
| Tumor Characteristics | Mammography Characteristics | US features | |||||
|---|---|---|---|---|---|---|---|
| Staging(Tumor Size)# [T] | Histologic type | Tumor Location | Position | Age at mammo (days) | Breast Density | Tumor Size (cm) | Tumor Size (cm) |
| 2 | 1 | L | L9 | NC | NC | 5 | 3 |
This table records the observation data from the Mammography and Ultra Sound images. So when we create FHIR resources for these EHR data we need to think about which MVP resources that we need.
Obviously, we need Patient resource to record the basic information of patient Linman. Then we also need two ImagingStudy resources for storing Mammography and Ultra Sound Dicom images. Finally, in terms of tumour size, location, position .etc data, we can store them into different Observation resources. And their reference relationship as shown in below graph:
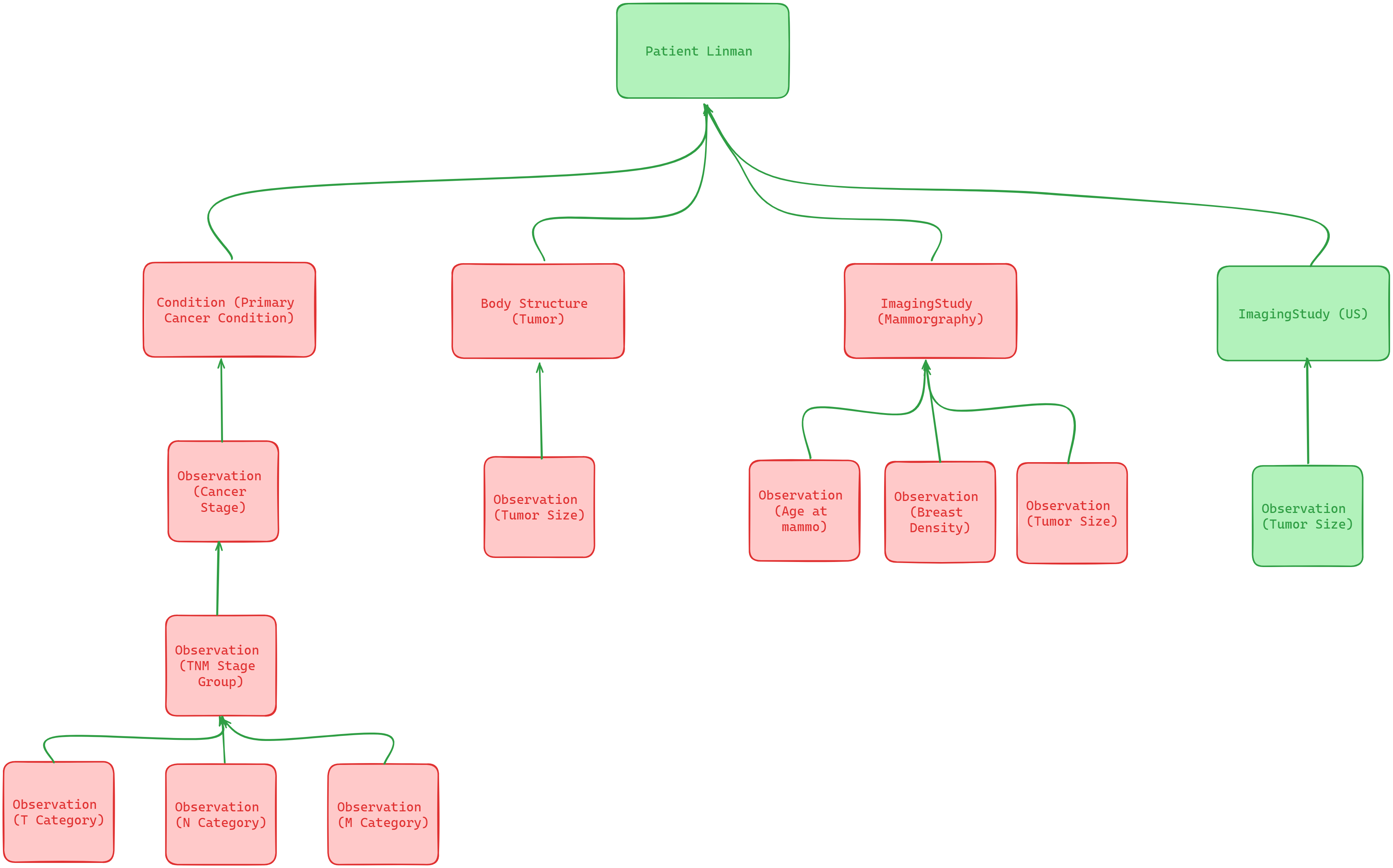
At this tutorial, we only show how to create Patient, Ultra sound ImagingStudy and tumour size Observation resource. As for others, you can follow by the same logic to create them in your database.
Setup environment
- Same to Tutorial 02
Introduction
Import library
At the beginning we need to import libraries fhirpy and os.
Also, we re-use the customise function pprint from Tutorial-02. We'll use it to display some Observation resource data structures.
import os
from fhirpy import AsyncFHIRClient
from pathlib import Path
import pydicom
from datetime import datetime
import uuidimport os
from fhirpy import AsyncFHIRClient
from pathlib import Path
import pydicom
from datetime import datetime
import uuidCreate an instance of connection
To load data from FHIR server we should instaniate FHIRClient class from fhirpy AsyncFHIRClient method.
Let's pass url and authorization arguments from environment.
client = AsyncFHIRClient(
url='http://localhost:8080/fhir/',
authorization='Bearer TOKEN',
)client = AsyncFHIRClient(
url='http://localhost:8080/fhir/',
authorization='Bearer TOKEN',
)Now, we are able to operate with FHIR resources using client.
Get patient 'Linman' from FHIR database
In the perviours tutorial we've already created patient Linman in the FHIR database. so we can get Linman by her name.
patientsResourceSearchSet = client.resources("Patient")
Linman = await patientsResourceSearchSet.search(name=['Linman']).first() patientsResourceSearchSet = client.resources("Patient")
Linman = await patientsResourceSearchSet.search(name=['Linman']).first()As a result, the dictionary stucture should looks like below:
Generate ImagingStudy Resource for Ultra sound Dicom images
The Dicom image metadata header tags you may interested in below tags. As for the information tags you can read DICOM Part 6 Data Dictionary.
(0010, 0010) Patient's Name(0010, 0020) Patient ID(0010, 0030) Patient's Birth Date(0010, 0040) Patient's Sex(0010, 1010) Patient's Age
As for create the ImagingResource, we also need to know which part of body does Linman take the UltraSound. As mentioned in senario, she took the UltraSound for her left breast. So we need to found the left breast code in SNOMED system:
{
"code": "73056007",
}{
"code": "73056007",
}Then, we can create the ImagingStudy resource for UltraSound Dicom image now
USImageStudyPath = "your dicom images' folder path"
patient = Linman
code = 'US'
display = 'Ultrasound'
bodySite = {
"system": "http://snomed.info/sct",
"code": "73056007",
"display": "Breast"
}
samples = sum(1 for item in USImageStudyPath.iterdir() if item.is_dir())
first_sample_path = next(iter(USImageStudyPath.glob('*')))
dicom_file = pydicom.dcmread(next(iter(first_sample_path.glob('*')), None))
study_uid = dicom_file[(0x0020, 0x000d)].value
try:
dicom_study_time = dicom_file[(0x0008, 0x0030)].value
started_time = datetime.strptime(dicom_study_time, "%H%M%S.%f").strftime("%Y-%m-%dT%H:%M:%S")
except:
started_time = datetime.now().strftime('%Y-%m-%dT%H:%M:%S')
try:
numberOfSeries = int(dicom_file[(0x0020, 0x1206)].value)
except:
numberOfSeries = sum(1 for item in USImageStudyPath.iterdir() if item.is_dir())
try:
numberOfInstances = int(dicom_file[(0x0020, 0x1208)].value)
except:
numberOfInstances = 0
for sample in USImageStudyPath.iterdir():
numberOfInstances += sum(1 for item in sample.iterdir() if item.is_file())
if patient is not None:
series = []
for sample in USImageStudyPath.iterdir():
if sample.is_file():
break
instances = []
first_file = next(iter(sample.glob('*')), None)
sample_dicom_file = pydicom.dcmread(first_file)
try:
numberOfSeriesInstances = int(sample_dicom_file[(0x0020, 0x1209)].value)
except:
numberOfSeriesInstances = sum(1 for item in sample.iterdir() if item.is_file())
for item in sample.iterdir():
if item.is_dir():
break
instance_dicom_file = pydicom.dcmread(item)
dicom_instance = {
"uid": instance_dicom_file[(0x0008, 0x0018)].value,
"number": instance_dicom_file[(0x0020, 0x0013)].value,
}
instances.append(dicom_instance)
series.append({
"uid": sample_dicom_file[(0x0020, 0x000e)].value,
"modality": {
"system": "http://dicom.nema.org/resources/ontology/DCM",
"code": code,
"display": display
},
"bodySite": bodySite,
"numberOfInstances": numberOfSeriesInstances,
"instance": instances
})
imagingResource = client.resource('ImagingStudy',
identifier=[{
"system": "urn:dicom:uid",
"value": f"urn:oid:{study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_study:uid",
"value": f"urn:uid:{USImageStudyPath.name + '-' + study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_dataset:uid",
"value": f"urn:uid:{str(USImageStudyPath.parent.parent.name) + '-' + str(uuid.uuid4())}"
},
],
status="available",
subject=patient.to_reference(),
started=started_time,
numberOfSeries=numberOfSeries,
numberOfInstances=numberOfInstances,
series=series
)
await imagingResource.save()USImageStudyPath = "your dicom images' folder path"
patient = Linman
code = 'US'
display = 'Ultrasound'
bodySite = {
"system": "http://snomed.info/sct",
"code": "73056007",
"display": "Breast"
}
samples = sum(1 for item in USImageStudyPath.iterdir() if item.is_dir())
first_sample_path = next(iter(USImageStudyPath.glob('*')))
dicom_file = pydicom.dcmread(next(iter(first_sample_path.glob('*')), None))
study_uid = dicom_file[(0x0020, 0x000d)].value
try:
dicom_study_time = dicom_file[(0x0008, 0x0030)].value
started_time = datetime.strptime(dicom_study_time, "%H%M%S.%f").strftime("%Y-%m-%dT%H:%M:%S")
except:
started_time = datetime.now().strftime('%Y-%m-%dT%H:%M:%S')
try:
numberOfSeries = int(dicom_file[(0x0020, 0x1206)].value)
except:
numberOfSeries = sum(1 for item in USImageStudyPath.iterdir() if item.is_dir())
try:
numberOfInstances = int(dicom_file[(0x0020, 0x1208)].value)
except:
numberOfInstances = 0
for sample in USImageStudyPath.iterdir():
numberOfInstances += sum(1 for item in sample.iterdir() if item.is_file())
if patient is not None:
series = []
for sample in USImageStudyPath.iterdir():
if sample.is_file():
break
instances = []
first_file = next(iter(sample.glob('*')), None)
sample_dicom_file = pydicom.dcmread(first_file)
try:
numberOfSeriesInstances = int(sample_dicom_file[(0x0020, 0x1209)].value)
except:
numberOfSeriesInstances = sum(1 for item in sample.iterdir() if item.is_file())
for item in sample.iterdir():
if item.is_dir():
break
instance_dicom_file = pydicom.dcmread(item)
dicom_instance = {
"uid": instance_dicom_file[(0x0008, 0x0018)].value,
"number": instance_dicom_file[(0x0020, 0x0013)].value,
}
instances.append(dicom_instance)
series.append({
"uid": sample_dicom_file[(0x0020, 0x000e)].value,
"modality": {
"system": "http://dicom.nema.org/resources/ontology/DCM",
"code": code,
"display": display
},
"bodySite": bodySite,
"numberOfInstances": numberOfSeriesInstances,
"instance": instances
})
imagingResource = client.resource('ImagingStudy',
identifier=[{
"system": "urn:dicom:uid",
"value": f"urn:oid:{study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_study:uid",
"value": f"urn:uid:{USImageStudyPath.name + '-' + study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_dataset:uid",
"value": f"urn:uid:{str(USImageStudyPath.parent.parent.name) + '-' + str(uuid.uuid4())}"
},
],
status="available",
subject=patient.to_reference(),
started=started_time,
numberOfSeries=numberOfSeries,
numberOfInstances=numberOfInstances,
series=series
)
await imagingResource.save()When we create the ImagingStudy resource, we also need to set the patient Linman reference to this UltraSound ImagingStudy resource.
Create Observation Resources
The Observation resource records the observation result from the UltraSound images. At this stage, from the table above we can see there is a tumour size result from UltraSound images.
Let's create the tumour size Observation resource for UltraSound ImagingStudy resource.
Remember, when we create the Observation resource, we need to find out the tumour size code under the loinc.org.
linman = await getPatient(client)
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference(), modality="US").fetch_all()
us_imagingstudy = imagingStudys[0]
us_observation = client.resource(
'Observation',
status='preliminary',
subject=linman.to_reference(),
focus=us_imagingstudy.to_reference(),
category=[{
'coding': [{
'system': 'http://terminology.hl7.org/CodeSystem/observation-category',
"code": "laboratory"
}]
}],
code={
'coding': [{
'system': 'http://loinc.org',
"code": "21889-1",
"display": "Size Tumor"
}]
},
effectiveDateTime="2018-04-01T00:00:00Z",
component=[
{
"code": {
"coding": [
{
"system": "http://loinc.org",
"code": "33728-7"
}
]
},
"valueQuantity": {
"value": 2.5,
"unit": "centimeters",
"system": "http://unitsofmeasure.org",
"code": "cm"
}
}
]
)
await us_observation.save()linman = await getPatient(client)
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference(), modality="US").fetch_all()
us_imagingstudy = imagingStudys[0]
us_observation = client.resource(
'Observation',
status='preliminary',
subject=linman.to_reference(),
focus=us_imagingstudy.to_reference(),
category=[{
'coding': [{
'system': 'http://terminology.hl7.org/CodeSystem/observation-category',
"code": "laboratory"
}]
}],
code={
'coding': [{
'system': 'http://loinc.org',
"code": "21889-1",
"display": "Size Tumor"
}]
},
effectiveDateTime="2018-04-01T00:00:00Z",
component=[
{
"code": {
"coding": [
{
"system": "http://loinc.org",
"code": "33728-7"
}
]
},
"valueQuantity": {
"value": 2.5,
"unit": "centimeters",
"system": "http://unitsofmeasure.org",
"code": "cm"
}
}
]
)
await us_observation.save()After we create the Observation resource for tumour size, we should take the patient Linman and her UltraSound ImagingStudy resources as references into subject and focus fields, respectively.
Search ImagingStudy Resource
Please view ImagingStudy Search Parameters and Observation Search Parameters to find out the parameters for search in you senario.
- Let's try search all studies/ImagingStudy Resources of a patient/volunteer
linmanaccross all SPARC Datasets. (Find all ImagingStudy Resources related tolinman)
linman = await getPatient(client)
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
# get all imagingStudy of patient linman
imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference()).fetch_all()
get all ultrasound imageStudy of patient linmanlinman = await getPatient(client)
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
# get all imagingStudy of patient linman
imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference()).fetch_all()
get all ultrasound imageStudy of patient linman- Let's try search the tumour size observation from the UltraSound image
imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference(), modality="US").fetch_all()
# get the linman's ultrasound observation resource
observation = await client.resources("Observation").search(patient=linman.to_reference(),
focus=imagingStudys[0].to_reference()).fetch_all()
print(observation)imagingStudys = await imagingStudyResourceSearchSet.search(patient=linman.to_reference(), modality="US").fetch_all()
# get the linman's ultrasound observation resource
observation = await client.resources("Observation").search(patient=linman.to_reference(),
focus=imagingStudys[0].to_reference()).fetch_all()
print(observation)Summary
In this tutorial the following topics were covered:
Patient,ImagingStudyandObservationresources.- How to use
pydicomto get dicom file tags information. - How to search
ImagingStudy Resourcefor specific identifier and speciality. - Search specific observation resource via ImagingStudy.