Data de-identification workflow in DigitalTWINS platform
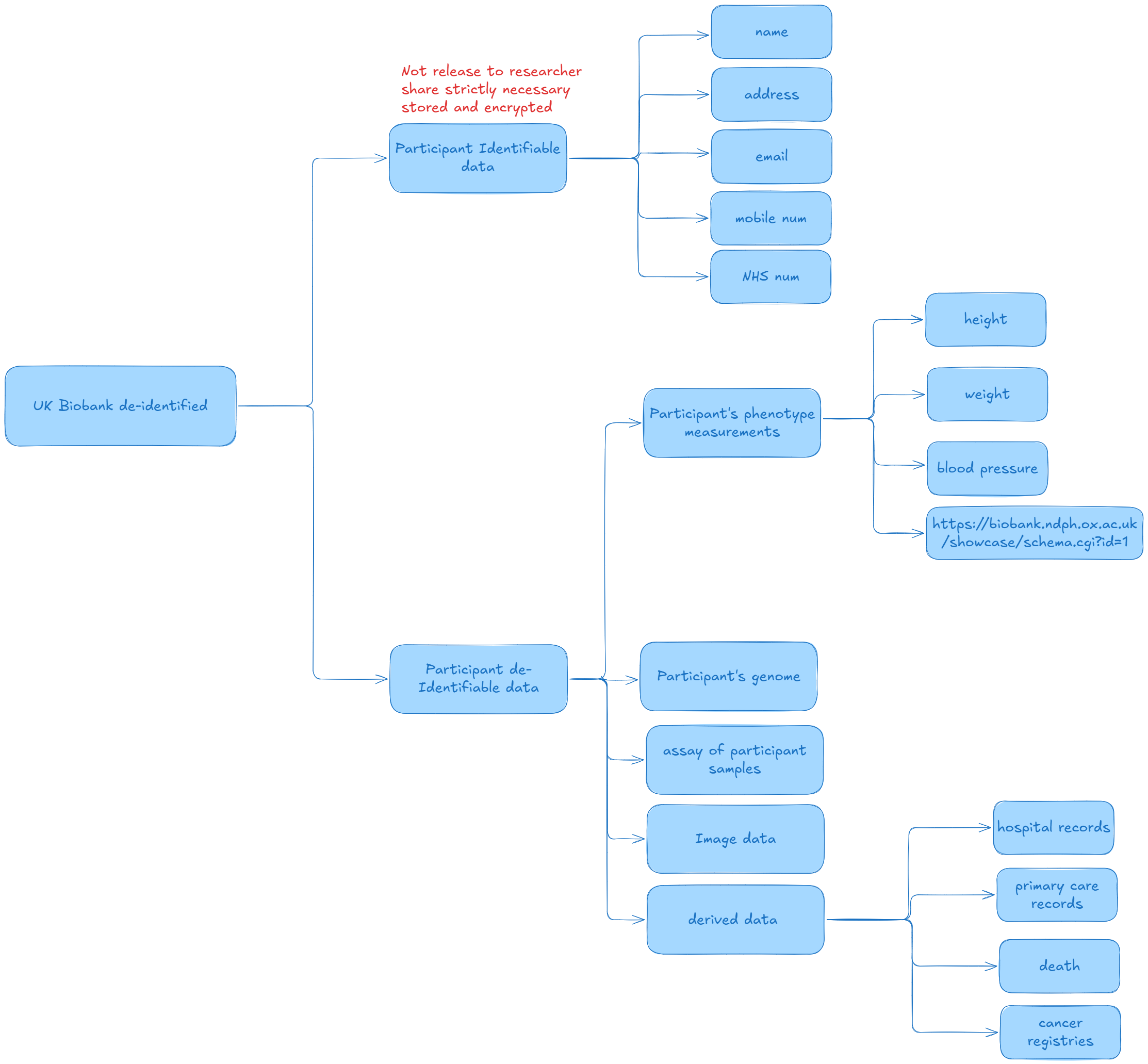
UK biobank de-identification
Purpose: Ensures participant data released to researchers is de-identified to prevent inadvertent re-identification, while complying with legal and ethical obligations.
Data Types:
Identifiable Data (not shared with researchers): Names, addresses, NHS numbers, contact details.
De-identified Data (shared with researchers): Phenotypes, genomic data, biomarkers, imaging, health records, and questionnaire responses.
Data Management:
Identifiable data is stored separately and encrypted, accessible only to authorized personnel.
A PID (unique identifier derived from NHS numbers) links datasets securely.
EID (project-specific encrypted identifier) replaces the PID in datasets shared with researchers.
HIPAA’s De-Identification Methods
Under the U.S. Health Insurance Portability and Accountability Act (HIPAA), two methods are recognized:
Safe Harbor Method:
Requires removal of 18 specific identifiers, including:
Names, addresses, phone numbers, email addresses.
Dates (e.g., birthdates, admission dates) except year.
Geographic subdivisions smaller than a state (e.g., full ZIP codes).
Social Security numbers, medical record numbers, health plan numbers.
Biometric identifiers (e.g., fingerprints), facial photos, unique device IDs.
Any remaining data is considered de-identified.
Expert Determination Method:
A qualified expert statistically certifies that the risk of re-identification is "very small."
Requires documentation of methods and results.
Key Differences
Date Handling:
HIPAA Safe Harbor requires dates to be reduced to year only.
UK Biobank truncates birthdates to month and year, which would not comply with HIPAA Safe Harbor.
Unique Identifiers:
- HIPAA prohibits sharing medical record numbers, while UK Biobank uses encrypted PIDs/EIDs (allowed under Expert Determination if risk is minimized).
Genetic Data:
- HIPAA does not explicitly address genomic data risks, whereas UK Biobank acknowledges genetic re-identification risks but deems them low.
Compliance Framework:
- HIPAA is legally mandated for U.S. entities; UK Biobank follows GDPR and contractual obligations (e.g., MTAs).
EUCanSHare
EUCanSHare, a project focused on cardiovascular data sharing across Europe, employs a comprehensive strategy to handle de-identified data, balancing research utility with privacy protection. Here's a structured overview of their approach:
De-identification Techniques:
Pseudonymization: Primary method where direct identifiers (e.g., names, IDs) are replaced with codes. A secure, separate key is maintained by a trusted third party or data custodian, allowing re-identification only under strict conditions.
Anonymization: Pursued where feasible, ensuring data cannot be linked back to individuals (e.g., aggregation, removal of indirect identifiers). This aligns with GDPR standards for irreversible anonymization.
Regulatory Compliance:
GDPR Adherence: Ensures all processes meet EU data protection standards. Pseudonymized data is treated as personal data, requiring safeguards like access controls and encryption.
Ethical and Legal Frameworks: Collaborates with ethics boards to ensure compliance with regional regulations and study-specific consent terms.
Data Governance and Access:
Tiered Access Model:
Open Access: Fully anonymized datasets available publicly.
Controlled Access: Pseudonymized data accessible via secure platforms, requiring ethical approval and data use agreements.
Data Use Agreements: Prohibit re-identification attempts and mandate secure handling.
Technical Safeguards:
Secure Infrastructure: Uses encrypted storage and transfer (e.g., HTTPS, VPNs) on platforms like the European Genome-Phenome Archive (EGA).
Metadata Management: Removes or obscures indirect identifiers (e.g., dates, rare diagnoses) in metadata to prevent linkage attacks.
Operational Protocols:
Centralized De-identification: Standardized processes applied at data hubs to ensure consistency across diverse sources (cohorts, registries).
Provenance Tracking: Maintains records of data transformations to preserve integrity without compromising privacy.
Ethical and Training Measures:
Informed Consent: Ensures participant consent covers de-identified data sharing, with studies audited for compliance.
Researcher Training: Provides guidelines on ethical data handling, GDPR, and de-identification best practices.
Risk Mitigation:
Regular Audits: Monitors data access and usage to prevent breaches.
Re-identification Risk Assessments: Evaluates datasets for residual risks, applying additional anonymization if needed.
De-identifying FHIR Patient Data with EUCanSHare
Scenario - FHIR data
We have FHIR Patient resources stored on your server. We want to share de-identified data for research via EUCanSHare while complying with privacy standards.
Identify Direct and Indirect Identifiers:
Direct identifiers (must be pseudonymized or removed):
Patient.id (internal system ID)
Patient.identifier (e.g., national ID, medical record number)
Patient.name
Patient.telecom (email/phone)
Patient.address
Patient.birthDate (if precise)
Indirect identifiers (may require generalization/removal):
Patient.gender
Patient.birthDate (year only)
Rare conditions (e.g., in Patient.extension or linked resources).
Apply De-identification Techniques:
- Original FHIR Patient Resource:
json{ "resourceType": "Patient", "id": "12345", "identifier": [{ "system": "urn:oid:2.16.840.1.113883.4.1", "value": "SSN-987-65-4321" }], "name": [{ "given": ["John"], "family": "Doe" }], "telecom": [{ "system": "phone", "value": "+1-555-123-4567" }], "gender": "male", "birthDate": "1985-07-15", "address": [{ "city": "Amsterdam", "country": "Netherlands" }] }{ "resourceType": "Patient", "id": "12345", "identifier": [{ "system": "urn:oid:2.16.840.1.113883.4.1", "value": "SSN-987-65-4321" }], "name": [{ "given": ["John"], "family": "Doe" }], "telecom": [{ "system": "phone", "value": "+1-555-123-4567" }], "gender": "male", "birthDate": "1985-07-15", "address": [{ "city": "Amsterdam", "country": "Netherlands" }] }- De-identified FHIR Patient Resource:
json{ "resourceType": "Patient", "id": "pseud-9a8b7c6d", // Pseudonymized ID Patient.id: "12345" → "pseud-9a8b7c6d" (using a secure hashing algorithm like SHA-256 with a salt). "identifier": [{ "system": "urn:oid:2.16.840.1.113883.4.1", "value": "pseud-a1b2c3d4" // Pseudonymized SSN }], "gender": "male", "birthDate": "1985", // Generalized to year only "address": [{ "country": "Netherlands" // Retain country, remove city }] }{ "resourceType": "Patient", "id": "pseud-9a8b7c6d", // Pseudonymized ID Patient.id: "12345" → "pseud-9a8b7c6d" (using a secure hashing algorithm like SHA-256 with a salt). "identifier": [{ "system": "urn:oid:2.16.840.1.113883.4.1", "value": "pseud-a1b2c3d4" // Pseudonymized SSN }], "gender": "male", "birthDate": "1985", // Generalized to year only "address": [{ "country": "Netherlands" // Retain country, remove city }] }Code for de-identification via EUCanSHare
import json
import hashlib
import secrets
from datetime import datetime
# Generate a secure salt (store this securely in production!)
SALT = secrets.token_hex(16)
def pseudonymize(value: str) -> str:
"""Pseudonymize a value using SHA-256 with salt"""
if not value:
return ""
return hashlib.sha256(f"{SALT}{value}".encode()).hexdigest()[:12]
def generalize_birthdate(date_str: str) -> str:
"""Generalize birthdate to year only"""
try:
return datetime.strptime(date_str, "%Y-%m-%d").strftime("%Y")
except:
return "" # Handle invalid/missing dates
def deidentify_patient(patient_data: dict) -> dict:
"""Main de-identification function for FHIR Patient resources"""
deidentified = {"resourceType": "Patient"}
# Pseudonymize direct identifiers
deidentified["id"] = pseudonymize(patient_data.get("id", ""))
# Handle identifiers (e.g., SSN)
if "identifier" in patient_data:
deidentified["identifier"] = [{
"system": ident["system"],
"value": pseudonymize(ident["value"])
} for ident in patient_data["identifier"]]
# Remove name and telecom completely
if "name" in patient_data:
deidentified["name"] = [{"family": "[REDACTED]", "given": ["[REDACTED]"]}]
# Generalize birthdate
if "birthDate" in patient_data:
deidentified["birthDate"] = generalize_birthdate(patient_data["birthDate"])
# Generalize address (keep country only)
if "address" in patient_data:
deidentified["address"] = [{
"country": addr.get("country", "")
} for addr in patient_data["address"]]
# Keep gender (low-risk)
deidentified["gender"] = patient_data.get("gender", "unknown")
return deidentified
# Example Usage
if __name__ == "__main__":
# Original FHIR Patient data
original_patient = {
"resourceType": "Patient",
"id": "12345",
"identifier": [{
"system": "urn:oid:2.16.840.1.113883.4.1",
"value": "SSN-987-65-4321"
}],
"name": [{
"given": ["John"],
"family": "Doe"
}],
"telecom": [{
"system": "phone",
"value": "+1-555-123-4567"
}],
"gender": "male",
"birthDate": "1985-07-15",
"address": [{
"city": "Amsterdam",
"country": "Netherlands"
}]
}
# De-identify the patient
deidentified_patient = deidentify_patient(original_patient)
# Remove telecom completely
if "telecom" in deidentified_patient:
del deidentified_patient["telecom"]
print("De-identified Patient:")
print(json.dumps(deidentified_patient, indent=2))import json
import hashlib
import secrets
from datetime import datetime
# Generate a secure salt (store this securely in production!)
SALT = secrets.token_hex(16)
def pseudonymize(value: str) -> str:
"""Pseudonymize a value using SHA-256 with salt"""
if not value:
return ""
return hashlib.sha256(f"{SALT}{value}".encode()).hexdigest()[:12]
def generalize_birthdate(date_str: str) -> str:
"""Generalize birthdate to year only"""
try:
return datetime.strptime(date_str, "%Y-%m-%d").strftime("%Y")
except:
return "" # Handle invalid/missing dates
def deidentify_patient(patient_data: dict) -> dict:
"""Main de-identification function for FHIR Patient resources"""
deidentified = {"resourceType": "Patient"}
# Pseudonymize direct identifiers
deidentified["id"] = pseudonymize(patient_data.get("id", ""))
# Handle identifiers (e.g., SSN)
if "identifier" in patient_data:
deidentified["identifier"] = [{
"system": ident["system"],
"value": pseudonymize(ident["value"])
} for ident in patient_data["identifier"]]
# Remove name and telecom completely
if "name" in patient_data:
deidentified["name"] = [{"family": "[REDACTED]", "given": ["[REDACTED]"]}]
# Generalize birthdate
if "birthDate" in patient_data:
deidentified["birthDate"] = generalize_birthdate(patient_data["birthDate"])
# Generalize address (keep country only)
if "address" in patient_data:
deidentified["address"] = [{
"country": addr.get("country", "")
} for addr in patient_data["address"]]
# Keep gender (low-risk)
deidentified["gender"] = patient_data.get("gender", "unknown")
return deidentified
# Example Usage
if __name__ == "__main__":
# Original FHIR Patient data
original_patient = {
"resourceType": "Patient",
"id": "12345",
"identifier": [{
"system": "urn:oid:2.16.840.1.113883.4.1",
"value": "SSN-987-65-4321"
}],
"name": [{
"given": ["John"],
"family": "Doe"
}],
"telecom": [{
"system": "phone",
"value": "+1-555-123-4567"
}],
"gender": "male",
"birthDate": "1985-07-15",
"address": [{
"city": "Amsterdam",
"country": "Netherlands"
}]
}
# De-identify the patient
deidentified_patient = deidentify_patient(original_patient)
# Remove telecom completely
if "telecom" in deidentified_patient:
del deidentified_patient["telecom"]
print("De-identified Patient:")
print(json.dumps(deidentified_patient, indent=2))De-identifying Dicom Data with HIPAA
Scenario - Dicom data
Now our dataset includes a series of dicom images that we need to de-identify them.
We can us e below code to do the de-identification based on HIPAA approach.
"""
anonymisation of DICOM files based on HIPAA standard
primary packaged: dicomanonymizer: https://pypi.org/project/dicom-anonymizer/
"""
import json
import pydicom
# from dicomanonymizer import anonymize, anonymizeDICOMFile, anonymizeDataset
import dicomanonymizer as anonymizer
def anon(dcm, anon_id):
tags_replaced_by_anon_id = [
(0x0010, 0x0020), # Patient ID
(0x0010, 0x0010) # Patient Name
]
for tag in tags_replaced_by_anon_id:
dcm[tag].value = anon_id
return dcm
def replace_by_anon_id(dataset, tag):
element = dataset.get(tag)
if element is not None:
element.value = anon_id
if __name__ == '__main__':
input_dicom_file = "/path_to_input/original.dcm"
output_dicom_path = "/path_to_output/anonymised.dcm"
anon_id = "anon_0001"
dcm = pydicom.read_file(input_dicom_file)
print(dcm)
print("patient id: ", dcm[(0x0010, 0x0020)].value)
print("patient name: ", dcm[(0x0010, 0x0010)].value)
f = open("original.json", "w")
f.write(str(dcm))
f.close()
# set custom rules
extra_rules = {}
tags_to_keep = [
(0x0010, 0x1010), # Patient's Age
(0x0010, 0x1030), # Patient's Weight
(0x0010, 0x1020) # Patient's Size
]
for tag in tags_to_keep:
extra_rules[tag] = anonymizer.keep
# for tag in anonymizer.ALL_TAGS:
# extra_rules[tag] = anonymizer.keep
tags_replaced_by_anon_id = [
(0x0010, 0x0020), # Patient ID
(0x0010, 0x0010) # Patient Name
]
for tag in tags_replaced_by_anon_id:
extra_rules[tag] = replace_by_anon_id
anonymizer.anonymize(
input_dicom_file,
output_dicom_path,
extra_rules,
delete_private_tags=False,
)
print("--------------------------------------------------------------------------------------------------")
print("anonymised!")
dcm = pydicom.read_file(output_dicom_path)
print(dcm)
print("patient id: ", dcm[(0x0010, 0x0020)].value)
print("patient name: ", dcm[(0x0010, 0x0010)].value)
f = open("anonymised.json", "w")
f.write(str(dcm))
f.close()"""
anonymisation of DICOM files based on HIPAA standard
primary packaged: dicomanonymizer: https://pypi.org/project/dicom-anonymizer/
"""
import json
import pydicom
# from dicomanonymizer import anonymize, anonymizeDICOMFile, anonymizeDataset
import dicomanonymizer as anonymizer
def anon(dcm, anon_id):
tags_replaced_by_anon_id = [
(0x0010, 0x0020), # Patient ID
(0x0010, 0x0010) # Patient Name
]
for tag in tags_replaced_by_anon_id:
dcm[tag].value = anon_id
return dcm
def replace_by_anon_id(dataset, tag):
element = dataset.get(tag)
if element is not None:
element.value = anon_id
if __name__ == '__main__':
input_dicom_file = "/path_to_input/original.dcm"
output_dicom_path = "/path_to_output/anonymised.dcm"
anon_id = "anon_0001"
dcm = pydicom.read_file(input_dicom_file)
print(dcm)
print("patient id: ", dcm[(0x0010, 0x0020)].value)
print("patient name: ", dcm[(0x0010, 0x0010)].value)
f = open("original.json", "w")
f.write(str(dcm))
f.close()
# set custom rules
extra_rules = {}
tags_to_keep = [
(0x0010, 0x1010), # Patient's Age
(0x0010, 0x1030), # Patient's Weight
(0x0010, 0x1020) # Patient's Size
]
for tag in tags_to_keep:
extra_rules[tag] = anonymizer.keep
# for tag in anonymizer.ALL_TAGS:
# extra_rules[tag] = anonymizer.keep
tags_replaced_by_anon_id = [
(0x0010, 0x0020), # Patient ID
(0x0010, 0x0010) # Patient Name
]
for tag in tags_replaced_by_anon_id:
extra_rules[tag] = replace_by_anon_id
anonymizer.anonymize(
input_dicom_file,
output_dicom_path,
extra_rules,
delete_private_tags=False,
)
print("--------------------------------------------------------------------------------------------------")
print("anonymised!")
dcm = pydicom.read_file(output_dicom_path)
print(dcm)
print("patient id: ", dcm[(0x0010, 0x0020)].value)
print("patient name: ", dcm[(0x0010, 0x0010)].value)
f = open("anonymised.json", "w")
f.write(str(dcm))
f.close()Create SPARC dataset for de-identify data
Once we finish the FHIR or dicom data de-identification, we can use sparc-me to create the SPARC dataset.
import sparc_me as sm
save_dir = "./tmp/template/"
dataset = sm.Dataset()
dataset.set_path(save_dir)
dataset.create_empty_dataset(version='2.0.0')
subjects = []
samples = []
sample1 = sm.Sample()
sample1.add_path("./test_data/bids_data/sub-01/sequence1/")
sample1.add_path(["./test_data/sample2/raw/dummy_sam2.txt", "./test_data/sample1/raw/dummy_sam1.txt"])
samples.append(sample1)
sample2 = sm.Sample()
sample2.add_path("./test_data/bids_data/sub-01/sequence2/")
samples.append(sample2)
subject1 = sm.Subject()
subject1.add_samples(samples)
subjects.append(subject1)
dataset.add_subjects(subjects)
dataset.save()import sparc_me as sm
save_dir = "./tmp/template/"
dataset = sm.Dataset()
dataset.set_path(save_dir)
dataset.create_empty_dataset(version='2.0.0')
subjects = []
samples = []
sample1 = sm.Sample()
sample1.add_path("./test_data/bids_data/sub-01/sequence1/")
sample1.add_path(["./test_data/sample2/raw/dummy_sam2.txt", "./test_data/sample1/raw/dummy_sam1.txt"])
samples.append(sample1)
sample2 = sm.Sample()
sample2.add_path("./test_data/bids_data/sub-01/sequence2/")
samples.append(sample2)
subject1 = sm.Subject()
subject1.add_samples(samples)
subjects.append(subject1)
dataset.add_subjects(subjects)
dataset.save()For more information, you can read sparc-me documentation and tutorials
Upload dataset to DigitalTwins platform
We can use digitaltwins-api package to upload the dataset.
from digitaltwins import Uploader
from pathlib import Path
if __name__ == '__main__':
dataset_dir = Path(r"/path/to/dataset_dir")
uploader = Uploader(Path(r"/path/to/configs.ini"))
uploader.upload(dataset_dir=dataset_dir)
print("done")from digitaltwins import Uploader
from pathlib import Path
if __name__ == '__main__':
dataset_dir = Path(r"/path/to/dataset_dir")
uploader = Uploader(Path(r"/path/to/configs.ini"))
uploader.upload(dataset_dir=dataset_dir)
print("done")For more information, you can read the digitaltwins-api documentation