Tutorial - FHIR ImagingStudy Resource
Tutorial code on GitHub hapi-py-fhir-tutorials -- imagingStudyOperator.py.
Currently, it is a private repository, will open source later...
Background
Scenario
Volunteers Aniyah and Linman recently had breast and heart MRIs taken at the Akron Hospital.The researcher at BioResearch Institute stored the collected MRI data in SPARC SDS datasets. As shown in the figure below, the researcher stored Aniyah and Linman's breast MRIs under the primary folder in the breast dataset, respectively.
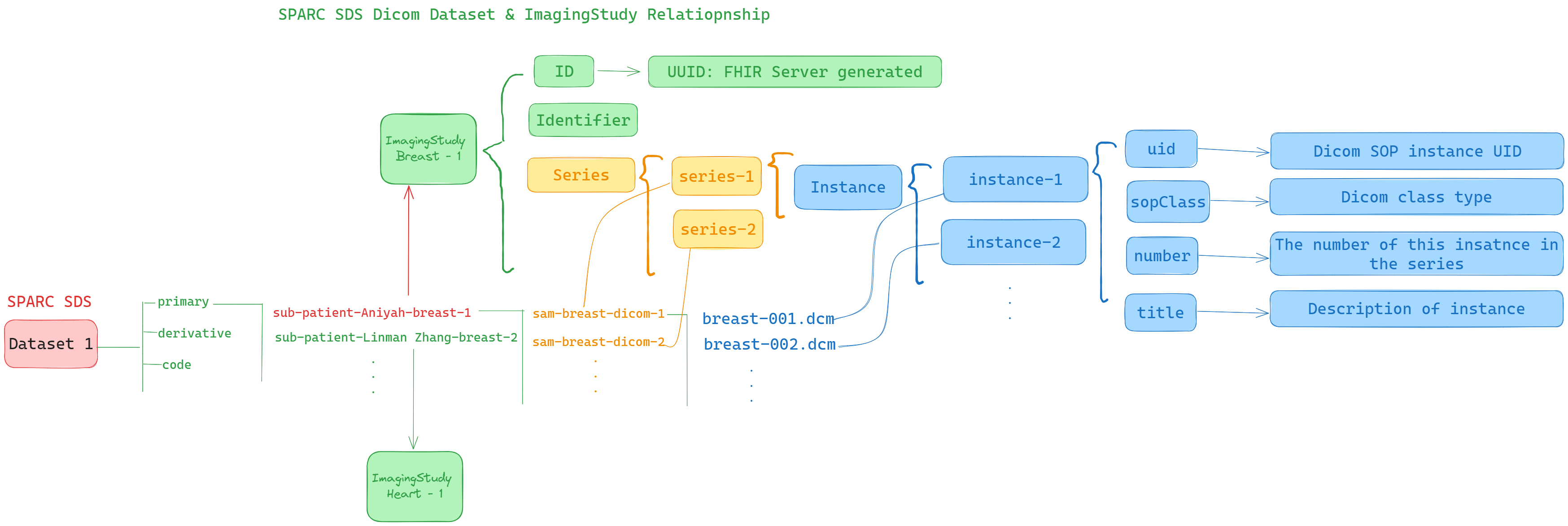
This figure shows the correspondence between the SPARC SDS dataset and the FHIR ImagingStudy Resource. From the figure we can see that each subject/study under the primary folder corresponds to an ImagingStudy object. The sample folder under each subject folder corresponds to the elements of the Series array of the ImagingStudy object. More over, the dicom files under each sample folder are stored one by one in the ImagingStudy.Series.Instance array.
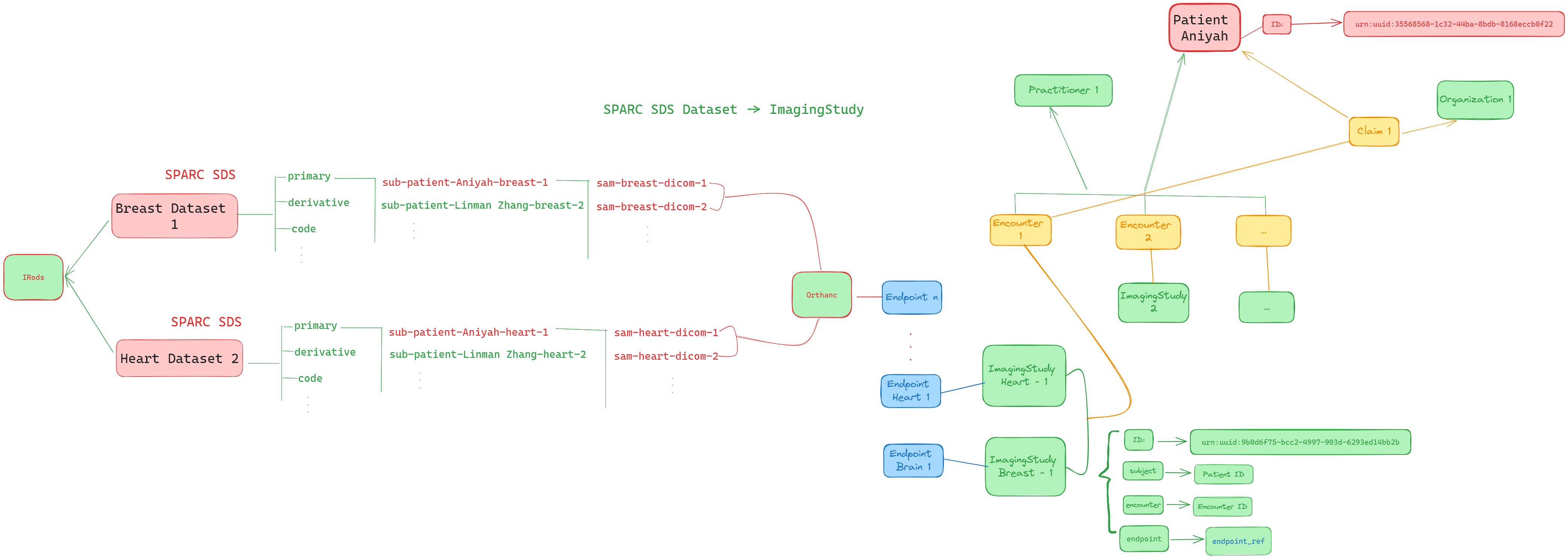
After we understand the relationship between SPARC SDS dataset and FHIR ImagingStudy Resource, now we can build the FHIR ImagingStudy resources for Aniyah and Linman. Below image shows how we create the SPARC SDS dataset and FHIR resources. From image we can understand, the Aniyah and Linman's breast and heart MRIs are stored in sparc-breast-dataset and sparc-heart-dataset. Then we can based on the diagram to convert the data to FHIR ImagingStudy resources.
Setup environment
- Same to Tutorial 02
Introduction
Import library
At the beginning we need to import libraries fhirpy and os.
Also, we re-use the customise function pprint from Tutorial-02. We'll use it to display some Observation resource data structures.
import os
from fhirpy import AsyncFHIRClient
from pathlib import Path
import pydicom
from datetime import datetime
import uuidimport os
from fhirpy import AsyncFHIRClient
from pathlib import Path
import pydicom
from datetime import datetime
import uuidCreate an instance of connection
To load data from FHIR server we should instaniate FHIRClient class from fhirpy AsyncFHIRClient method.
Let's pass url and authorization arguments from environment.
client = AsyncFHIRClient(
url='http://localhost:8080/fhir/',
authorization='Bearer TOKEN',
)client = AsyncFHIRClient(
url='http://localhost:8080/fhir/',
authorization='Bearer TOKEN',
)Now, we are able to operate with FHIR resources using client.
Read and Perpare SPARC SDS dataset Dicom files structure
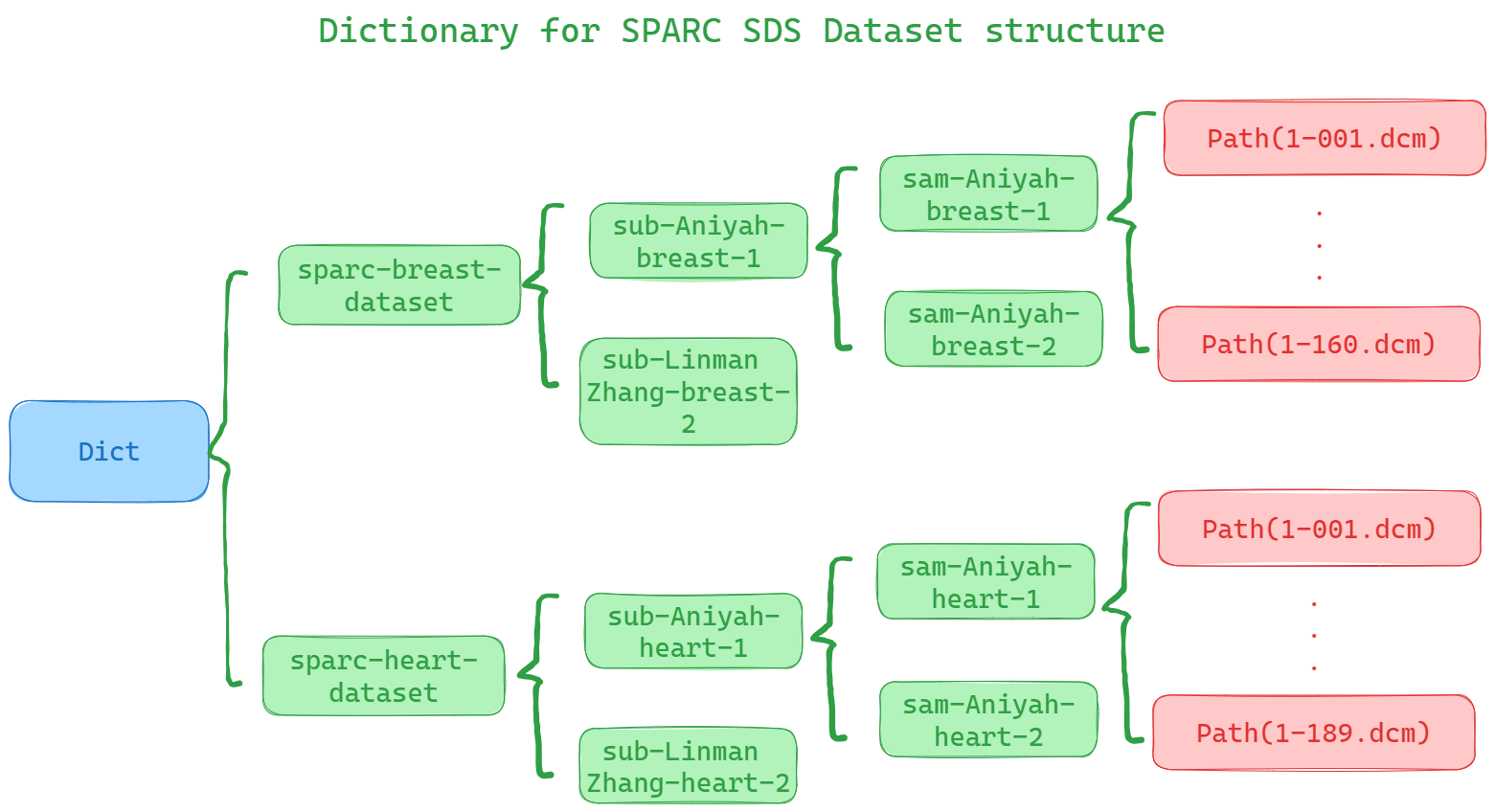
For easily to create a ImagingStudy resource, we can read the whole SPARC datasets structure into a dictionary. This is in order to use pydicom to read each dicom file header informations.
sparc_fhir_structure = {}
sparc_data_paths = [Path("./sparc_fhir_breast_dataset/primary/"), Path("./sparc_fhir_heart_dataset/primary/")]
for sparc_data_path in sparc_data_paths:
sparc_fhir_structure[sparc_data_path.parent] = {}
for study in sparc_data_path.iterdir():
if study.is_dir():
sparc_fhir_structure[sparc_data_path.parent][study.name] = {}
for series in study.iterdir():
if series.is_dir():
sparc_fhir_structure[sparc_data_path.parent][study.name][series.name] = list(
series.glob('*.dcm'))sparc_fhir_structure = {}
sparc_data_paths = [Path("./sparc_fhir_breast_dataset/primary/"), Path("./sparc_fhir_heart_dataset/primary/")]
for sparc_data_path in sparc_data_paths:
sparc_fhir_structure[sparc_data_path.parent] = {}
for study in sparc_data_path.iterdir():
if study.is_dir():
sparc_fhir_structure[sparc_data_path.parent][study.name] = {}
for series in study.iterdir():
if series.is_dir():
sparc_fhir_structure[sparc_data_path.parent][study.name][series.name] = list(
series.glob('*.dcm'))As a result, the dictionary stucture should looks like below:
Create Patients
Here, for this tutorial we can create two dummy patients for test. In a real development, we need to read the dicom header to find out the patient's information then based on that information to create the Patient Resource. As for the information tags you can read DICOM Part 6 Data Dictionary, here provides some examples:
(0010, 0010) Patient's Name(0010, 0020) Patient ID(0010, 0030) Patient's Birth Date(0010, 0040) Patient's Sex(0010, 1010) Patient's Age
The Patient here is used for reference in ImagingStudy resource.
Aniyah = client.resource('Patient',
name=[
{
'given': ['Aniyah'],
'family': '',
'use': 'official',
'prefix': ['Mr. '],
}
],
gender="female",
brithDate='1995-09-22'
)
Linman = client.resource('Patient',
name=[
{
'given': ['Linman'],
'family': 'Zhang',
'use': 'official',
'prefix': ['Mr. '],
}
],
gender="female",
brithDate='1993-08-15'
)
await Aniyah.save()
await Linman.save()Aniyah = client.resource('Patient',
name=[
{
'given': ['Aniyah'],
'family': '',
'use': 'official',
'prefix': ['Mr. '],
}
],
gender="female",
brithDate='1995-09-22'
)
Linman = client.resource('Patient',
name=[
{
'given': ['Linman'],
'family': 'Zhang',
'use': 'official',
'prefix': ['Mr. '],
}
],
gender="female",
brithDate='1993-08-15'
)
await Aniyah.save()
await Linman.save()Create ImagingStudy Resources
ImagingStudy resource provides mappings of its elements to DICOM attributes. DICOM attributes are identified by a 32-bit tag, presented in canonical form as two four-digit hexadecimal values within parentheses and separated by a comma, e.g. (0008, 103E). The name and value representation (data type) of each attribute can be found in DICOM Part 6 Data Dictionary. The use of the attributes in the context of information objects, including detailed description of use, can be found in DICOM Part 3 Information Object Definitions. Attributes used in the DICOM query information models, such as Number of Instances in Study, can be found in DICOM Part 4 Annex.
DICOM Series Instance UID and SOP Instance UID use the id datatype, and are encoded directly. For example, an image with SOP Instance UID of 2.16.124.113543.1154777499.30246.19789.3503430045.1.1 is encoded in ImagingStudy.series.instance.uid as 2.16.124.113543.1154777499.30246.19789.3503430045.1.1.
The ImagingStudy's DICOM Study Instance UID is encoded in the ImagingStudy.identifier element, which is of the Identifier datatype. When encoding a DICOM UID in an Identifier datatype, use the Identifier system of "urn:dicom:uid", and prefix the UID calue with "urn:oid:". Therefore, an ImagingStudy with DICOM Study Instance UID of 2.16.124.113543.1154777499.30246.19789.3503430046 is encoded as:
{
"identifier": {
"system": "urn:dicom:uid",
"value": "urn:oid:2.16.124.113543.1154777499.30246.19789.3503430046"
}
}{
"identifier": {
"system": "urn:dicom:uid",
"value": "urn:oid:2.16.124.113543.1154777499.30246.19789.3503430046"
}
}Here are some useful dicom tags for building ImagingStudy Resource:
- (0020, 000d) Study Instance UID
- (0020, 000e) Series Instance UID
- (0008, 0018) SOP Instance UID
- (0020, 0013) Instance Number
- (0008, 0030) Study Time
- (0020,1208) Number of Study Related Instances
- (0020,1206) Number of Study Related Series
- (0020,1209) Number of Series Related Instances
- (0018, 0010) Contrast/Bolus Agent
- (0018, 0015) Body Part Examined
Let's create a ImagingStudy for Aniyah's breast study
Get the study/samples for SPARC dictionary
- Get study/samples
Aniyah_breast_study = sparc_fhir_structure["sparc_fhir_breast_dataset"]["sub-Aniyah-breast-1"]Aniyah_breast_study = sparc_fhir_structure["sparc_fhir_breast_dataset"]["sub-Aniyah-breast-1"]Using pydicom to find header informations for ImagingStudy
Using pydicom to read the a series/sample dicom file header tags, we can get the Study Instance UID, Number of Study Related Instances, Number of Study Related Series, Study Time.
Because of de-identification dicom data, some tags will missing. So we need use try except to catch the error.
dicom_file = pydicom.dcmread(Aniyah_breast_study[0][0])
study_uid = dicom_file[(0x0020, 0x000d)].value
try:
dicom_study_time = dicom_file[(0x0008, 0x0030)].value
started_time = datetime.strptime(dicom_study_time, "%H%M%S.%f").strftime("%Y-%m-%dT%H:%M:%S")
except:
started_time = datetime.now().strftime('%Y-%m-%dT%H:%M:%S')
try:
numberOfSeries = int(dicom_file[(0x0020, 0x1206)].value)
except:
numberOfSeries = len(Aniyah_breast_study)
try:
numberOfInstances = int(dicom_file[(0x0020, 0x1208)].value)
except:
numberOfInstances = 0
for sample in Aniyah_breast_study:
numberOfInstances += len(sample)dicom_file = pydicom.dcmread(Aniyah_breast_study[0][0])
study_uid = dicom_file[(0x0020, 0x000d)].value
try:
dicom_study_time = dicom_file[(0x0008, 0x0030)].value
started_time = datetime.strptime(dicom_study_time, "%H%M%S.%f").strftime("%Y-%m-%dT%H:%M:%S")
except:
started_time = datetime.now().strftime('%Y-%m-%dT%H:%M:%S')
try:
numberOfSeries = int(dicom_file[(0x0020, 0x1206)].value)
except:
numberOfSeries = len(Aniyah_breast_study)
try:
numberOfInstances = int(dicom_file[(0x0020, 0x1208)].value)
except:
numberOfInstances = 0
for sample in Aniyah_breast_study:
numberOfInstances += len(sample)Create Series and Instance Array of ImagingStudy
Because we have a sparc dictionary stucture, we can easily to use for loop to create the Series and Instance Array. In here we need to use Series Instance UID, SOP Instance UID, Instance Number, Number of Series Related Instances tags.
As for the series modality code, we can find it in dicom website.
series = []
for sample in Aniyah_breast_study:
instances = []
sample_dicom_file = pydicom.dcmread(sample[0])
try:
numberOfSeriesInstances = int(sample_dicom_file[(0x0020, 0x1209)].value)
except:
numberOfSeriesInstances = len(sample)
for item in sample:
instance_dicom_file = pydicom.dcmread(item)
dicom_instance = {
"uid": instance_dicom_file[(0x0008, 0x0018)].value,
"number": instance_dicom_file[(0x0020, 0x0013)].value,
}
instances.append(dicom_instance)
series.append({
"uid": sample_dicom_file[(0x0020, 0x000e)].value,
"modality": {
"system": "http://dicom.nema.org/resources/ontology/DCM",
"code": "MR"
},
"numberOfInstances": numberOfSeriesInstances,
"instance": instances
})
series = []
for sample in Aniyah_breast_study:
instances = []
sample_dicom_file = pydicom.dcmread(sample[0])
try:
numberOfSeriesInstances = int(sample_dicom_file[(0x0020, 0x1209)].value)
except:
numberOfSeriesInstances = len(sample)
for item in sample:
instance_dicom_file = pydicom.dcmread(item)
dicom_instance = {
"uid": instance_dicom_file[(0x0008, 0x0018)].value,
"number": instance_dicom_file[(0x0020, 0x0013)].value,
}
instances.append(dicom_instance)
series.append({
"uid": sample_dicom_file[(0x0020, 0x000e)].value,
"modality": {
"system": "http://dicom.nema.org/resources/ontology/DCM",
"code": "MR"
},
"numberOfInstances": numberOfSeriesInstances,
"instance": instances
})Create a Aniyah's breast ImagingStudy
As we discussed the identifier before, we need use "urn:dicom:uid" system to store the study uid as the identifier value. Here, we can create two customise identifier for easily mapping the SPARC SDS dataset and FHIR ImagingStudy resource structure. So we can create two local identifier for the SPARC SDS system: urn:sparc_study:uid and urn:sparc_dataset:uid.
urn:sparc_study:uid: is used to store the unique subject/study folder name in SPARC SDS dataset to ImagingStudy identifier.urn:sparc_dataset:uid: is used to store the unique sparc dataset name to ImagingStudy identifier.
imagingResource = client.resource('ImagingStudy',
identifier=[{
"system": "urn:dicom:uid",
"value": f"urn:oid:{study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_study:uid",
"value": f"urn:uid:{"sub-Aniyah-breast-1" + '-' + study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_dataset:uid",
"value": f"urn:uid:{"sparc_fhir_breast_dataset" + '-' + str(uuid.uuid4())}"
},
],
status="available",
subject=Aniyah.to_reference(),
started=started_time,
numberOfSeries=numberOfSeries,
numberOfInstances=numberOfInstances,
series=series
)
await imagingResource.save()imagingResource = client.resource('ImagingStudy',
identifier=[{
"system": "urn:dicom:uid",
"value": f"urn:oid:{study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_study:uid",
"value": f"urn:uid:{"sub-Aniyah-breast-1" + '-' + study_uid}"
},
{
"use": 'temp',
"system": "urn:sparc_dataset:uid",
"value": f"urn:uid:{"sparc_fhir_breast_dataset" + '-' + str(uuid.uuid4())}"
},
],
status="available",
subject=Aniyah.to_reference(),
started=started_time,
numberOfSeries=numberOfSeries,
numberOfInstances=numberOfInstances,
series=series
)
await imagingResource.save()Update ImagingStudy Resource
Under creating ImagingStudy part, we forgot to create bodySite in each series. We can find the code in SNOMED CT.
- BodySite: The anatomic structures examined. See DICOM Part 16 Annex L for DICOM to SNOMED-CT mappings. The bodySite may indicate the laterality of body part imaged; if so, it shall be consistent with any content of
ImagingStudy.series.laterality.
The code for breast and heart:
Breast:76752008Heart:80891009
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
count = await imagingStudyResourceSearchSet.count()
# Get all imagingstudy resources
imagingStudys = await imagingStudyResourceSearchSet.limit(count).fetch()
# find a patient in which datasets
for imagingstudy in imagingStudys:
if "identifier" in imagingstudy:
for identifier in imagingstudy['identifier']:
if identifier.get('system') == 'urn:sparc_dataset:uid':
for index, series in enumerate(imagingstudy["series"]):
if "breast" in identifier.get('value'):
imagingstudy["series"][index]["bodySite"] = {
"system": "http://snomed.info/sct",
"code": "76752008",
"display": "Breast"
}
elif "heart" in identifier.get('value'):
imagingstudy["series"][index]["bodySite"] = {
"system": "http://snomed.info/sct",
"code": "80891009",
"display": "Heart"
}
await imagingstudy.save()imagingStudyResourceSearchSet = client.resources('ImagingStudy')
count = await imagingStudyResourceSearchSet.count()
# Get all imagingstudy resources
imagingStudys = await imagingStudyResourceSearchSet.limit(count).fetch()
# find a patient in which datasets
for imagingstudy in imagingStudys:
if "identifier" in imagingstudy:
for identifier in imagingstudy['identifier']:
if identifier.get('system') == 'urn:sparc_dataset:uid':
for index, series in enumerate(imagingstudy["series"]):
if "breast" in identifier.get('value'):
imagingstudy["series"][index]["bodySite"] = {
"system": "http://snomed.info/sct",
"code": "76752008",
"display": "Breast"
}
elif "heart" in identifier.get('value'):
imagingstudy["series"][index]["bodySite"] = {
"system": "http://snomed.info/sct",
"code": "80891009",
"display": "Heart"
}
await imagingstudy.save()Search ImagingStudy Resource
Please view ImagingStudy Search Parameters to find out the parameters for search in you senario.
- Let's try search all studies/ImagingStudy Resources of a patient/volunteer
Aniyahaccross all SPARC Datasets. (Find all ImagingStudy Resources related toAniyah)
patientsResourceSearchSet = client.resources("Patient")
Aniyah = await patientsResourceSearchSet.search(name=['Aniyah']).first()
# find all studies of a patient cross all datasets
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
imagingStudys = await imagingStudyResourceSearchSet.search(patient=Aniyah.to_reference()).fetch_all()
pprint(imagingStudys)patientsResourceSearchSet = client.resources("Patient")
Aniyah = await patientsResourceSearchSet.search(name=['Aniyah']).first()
# find all studies of a patient cross all datasets
imagingStudyResourceSearchSet = client.resources('ImagingStudy')
imagingStudys = await imagingStudyResourceSearchSet.search(patient=Aniyah.to_reference()).fetch_all()
pprint(imagingStudys)- Find
Aniyah'sImagingStudy Resources in which SPARC SDS dataset
# find a patient in which datasets
for imagingstudy in imagingStudys:
for identifier in imagingstudy['identifier']:
if identifier.get('system') == 'urn:sparc_dataset:uid':
print(identifier.get('value')) # find a patient in which datasets
for imagingstudy in imagingStudys:
for identifier in imagingstudy['identifier']:
if identifier.get('system') == 'urn:sparc_dataset:uid':
print(identifier.get('value'))- Find all
BreastImagingStudy resources viabodysite.
# find all breast ImagingStudy resources
imagingstudys = await client.resources('ImagingStudy').search(bodysite="76752008").fetch_all()
print(imagingstudys)# find all breast ImagingStudy resources
imagingstudys = await client.resources('ImagingStudy').search(bodysite="76752008").fetch_all()
print(imagingstudys)- Find all
HeartImagingStudy resources viabodysite.
# find all heart ImagingStudy resources
imagingstudys = await client.resources('ImagingStudy').search(bodysite="80891009").fetch_all()
print(imagingstudys)# find all heart ImagingStudy resources
imagingstudys = await client.resources('ImagingStudy').search(bodysite="80891009").fetch_all()
print(imagingstudys)- Find a specific ImagingStudy resource if you have its identifier value
specific_ImagingStudy = await imagingStudyResourceSearchSet.search(identifier='urn:uid:sparc_fhir_heart_dataset-9c3319e9-38f0-42fc-9b41-28a1be44f72e').fetch()
print(specific_ImagingStudy)specific_ImagingStudy = await imagingStudyResourceSearchSet.search(identifier='urn:uid:sparc_fhir_heart_dataset-9c3319e9-38f0-42fc-9b41-28a1be44f72e').fetch()
print(specific_ImagingStudy)Summary
In this tutorial the following topics were covered:
ImagingStudyresources.- SPARC SDS dataset and ImagingStudy Resource relationship.
- How to use
pydicomto get dicom file tags information. - How to create a ImagingStudy Resource.
- How to update a ImagingStudy Resource.
- How to search
ImagingStudy Resourcefor specific identifier and speciality.