Guideline for SODA
Introduce
SODA (Software to Organize Data Automatically) is a desktop (MacOS, Windows, and Ubuntu) GUI software for creating SPARC SDS dataset.
Perparation for SODA
- A
ORCID iDfor each dataset contributor, if you don't have you can register one on here, or you canAccess through your institutionto get the ID. - A
pennsieveaccount, if you don't have you can register one on here. - Download the SODA follow by the install guideline here. After you download the SODA and run it correctly, it will automatically ask you to download a pennsive plugin, then you can just click to download it. At last, you will be ask to login in your pennsieve account in SODA, then it is ready for you to start creating a dataset on SODA.
Metadata Files and Category folders in SPARC SDS Dataset
- Metadata files requirments:
- Mandatory high-level metadata files:
submission,dataset_description,README - Dataset-specific high-level metadata files:
subjects,samples,CHANGES - Code-related metadata files:
code_description,code_parameters
- Mandatory high-level metadata files:
- Guide structure, rules and examples for metadata files can be found in:
- Informations for each category folder data
Primary folder/data: refers to all data collected from subjects/samples (e.g, time series data, tabular data, clinical imaging data, genomic, metabolomic, or microscopy data). The data generally have been minimally processed so they are in a form ready for analysis.Source folder/data: refers to unaltered, raw files from an experiment. For example, this may include the "truly" raw k-space data for a Magnetic Resonance (MR) image that has not yet been reconstructed (in this case, the reconstructed DICOM or NIFTI files would be found within the primary folder).Derivative folder/data: is any data products from the original data (i.e, primary or source data). Derivative data includes data such as measurements from images, 3d reconstructions from serial sections, or file conversions.Code folder/data: Your code files in your dataset. When you copy and paste the code files in to code folder, please make sure you include a README file that describes the code and how to use it.Protocol folder/data: refers to supplementary folders and files to accompany the experimental protocols that must be submitted to protocols.io.Docs folder/data: refers to any data to be included in your dataset that does not belong to any of the other five categories (primary, source, derivative, code, or protocol data).
Implementation
SODA has two modes to create a dataset: End to End Curation and Free Form Curation. Based on the SODA official best practice, they recommend if you are create a new dataset may need to choose End to End mode, if you are planning to edit and making a few changes on exsiting dataset, they suggest to use Free Form Curation mode.
End to End Curation mode, it provide very detailed and step-by-step guideline in the software, when you create a dataset via this mode.- NOTE: the SPARC guidelines under this mode require all datasets to be uploaded to Pennsieve. So you must have a pennisve account, otherwise you cannot generate the dataset.
- Guide user to choose which metadata files and dataset folders they need in their dataset.
- If user already have metadata files, they can upload it directly, otherwise they can follow the guideline to create a new one.
- Make forms for each metadata file, and highlight the required elements with
*. - If some elements have relationship among different metadata files/forms, it will give a reminder to user or will automatically fill in each form. E.g, the
Fundingelement in submission metadata is related to theFundingelement in dataset_description metadata, so when we edit on dataset_description the SODA will give us a reminder about that. - Supports user to create folders in dataset structure folders. Assume we have
primary,derivative,docs,code, andsourcefolders in the dataset structure. We can very easily to achieve these things: - Create, edit and delete folders/files under these above folders. - Copy and Pastepool,SubjectorSamplefolders intoprimary,derivativeandsourcefolders. But the structure order should absolutely like: pool/subject/sample. And copy files into dataset samples folder.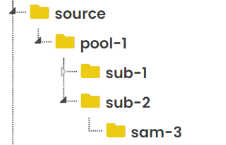 - It will automatically generate the manifest metadata files for each folders (
- It will automatically generate the manifest metadata files for each folders (primary,derivative,docs,code, andsource), if you've already copied files/folders into them.
Free Form Curation mode, it don't have too much detailed guideline details for user.- If you are familar with SPARC dataset and the requirements in each metadata files, you can create it in this way very fast.
- It allows (must) user to upload the metadata files, but would not automatically generate some elements for them. (subject_id, sample_id and pool_id, number of subjects/samples).
- Will automatically generate the manifest in each folder, if you upload foldes/files in that folder(primary/derivative/source).
- Also if you haven't upload any files in a folder or a metadata file, it would not generate that folder or metadata file for you in a dataset, even if them appears in dataset structure figure.
- Notice:
- It would not generate the upload files into dataset, you need to manually copy and paste into the generate dataset. It seems just use the upload files names to generate the manifest file in that folder.
- There is no validation guidelines when you upload metadata files into SODA. So you need to make sure all your metadata informations are correct.
- It won't automatically filled some common relational elements for you. E.g, the subject id and sample id in their
subjectsandsamplesmetadata files.
Create SPARC SDS dataset via SODA End to End Curation
- Create Dataset Name
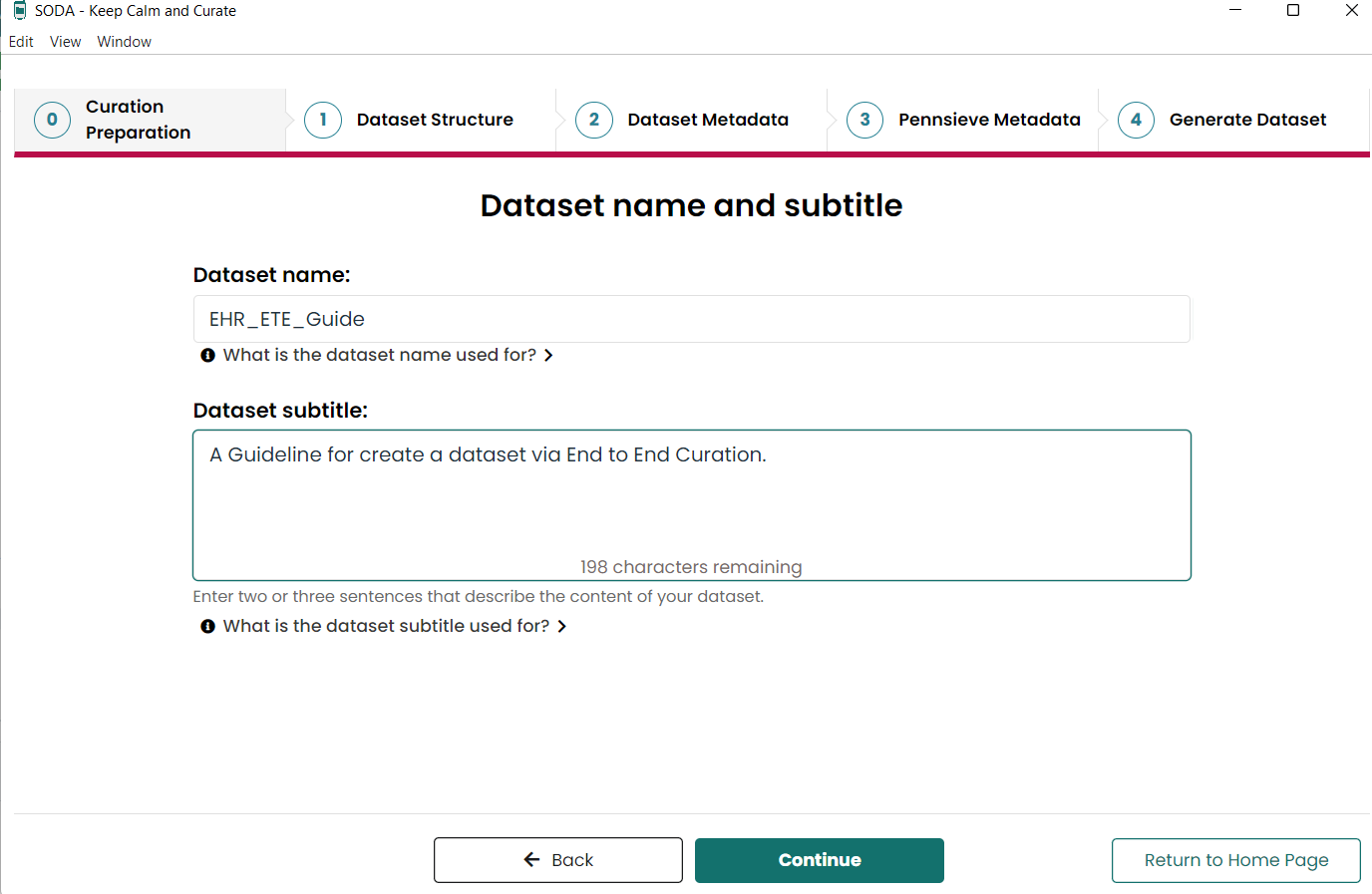
- Generate Dataset Structure
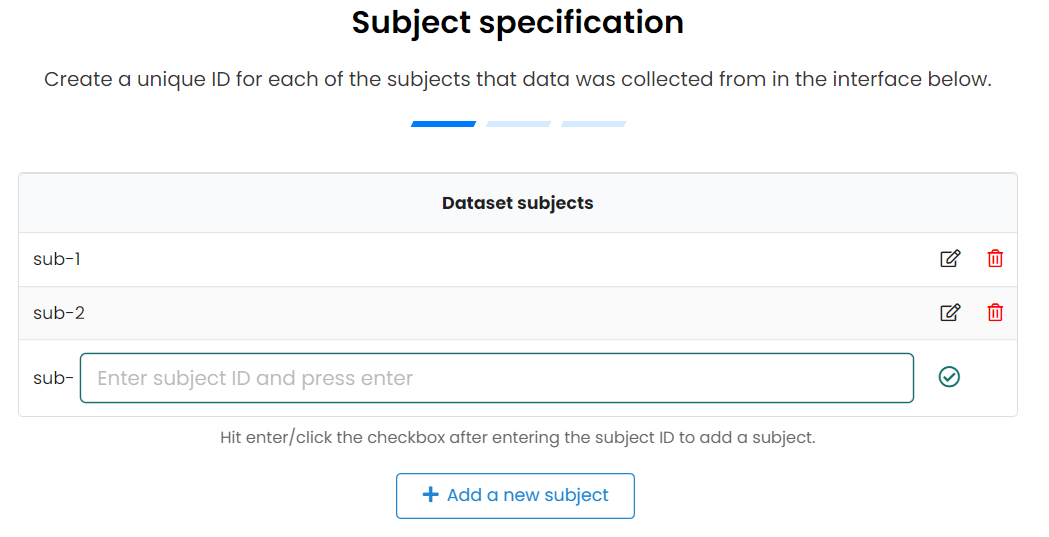
- To tell SODA how many subjects and samples you have
- Need to specify their unique name
- Can organise subjects into different pools
- Upload your data/files into different categories (primary, source, derivative, code, docs, or protocol) folder.
- NOTE: all empty folders will be deleted before generating the dataset since empty folders are not allowed in the SDS.
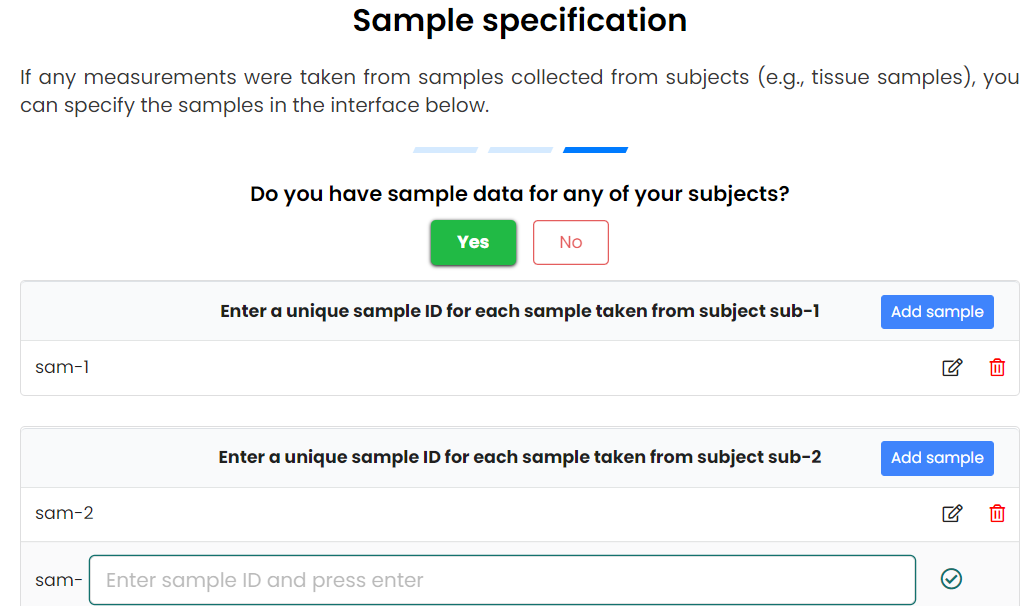
- Upload primary data for samples/subjects/pools
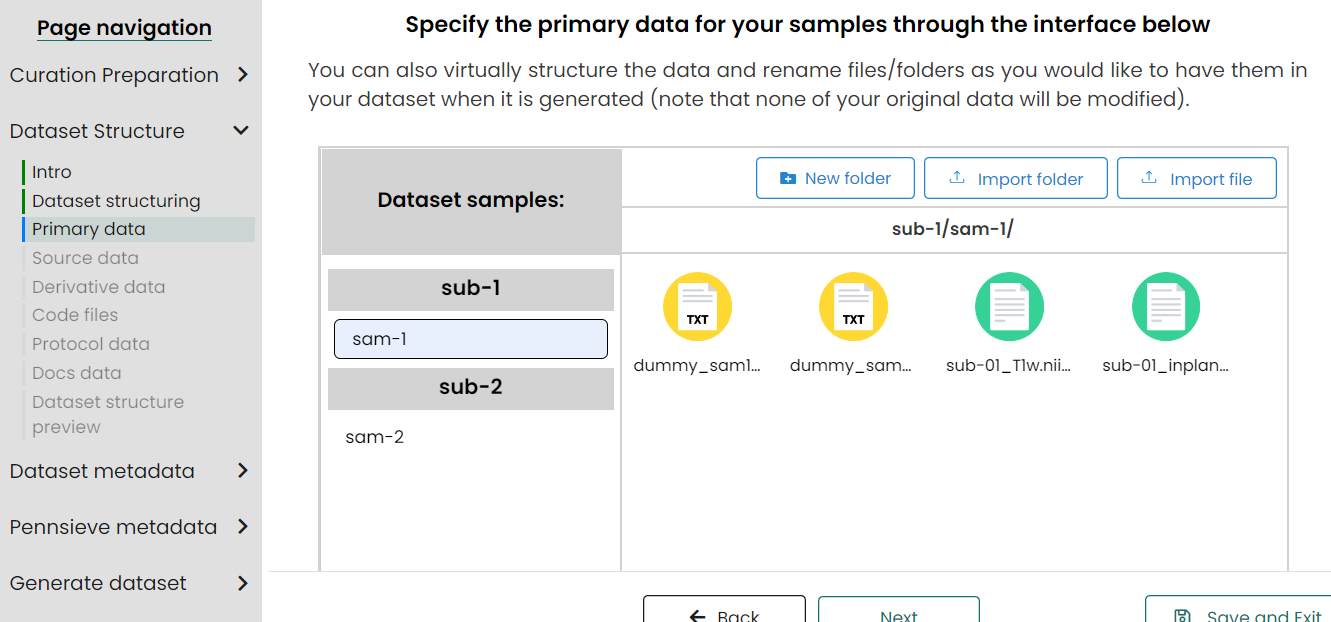
Upload
source, derivative, code, docs, or protocol dataare same steps as above.Edit Dataset Metadata
- Manifest The SODA will automatically generate manifest files in each category folders if they have files (not an empty folder). In these step you also can view and edit each manifest metadata file.
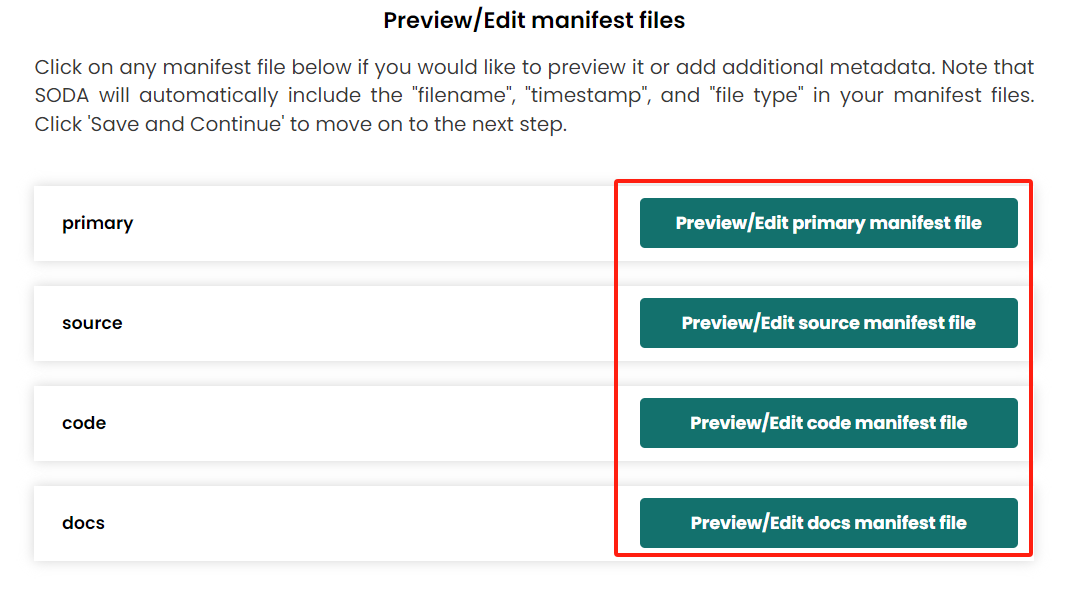
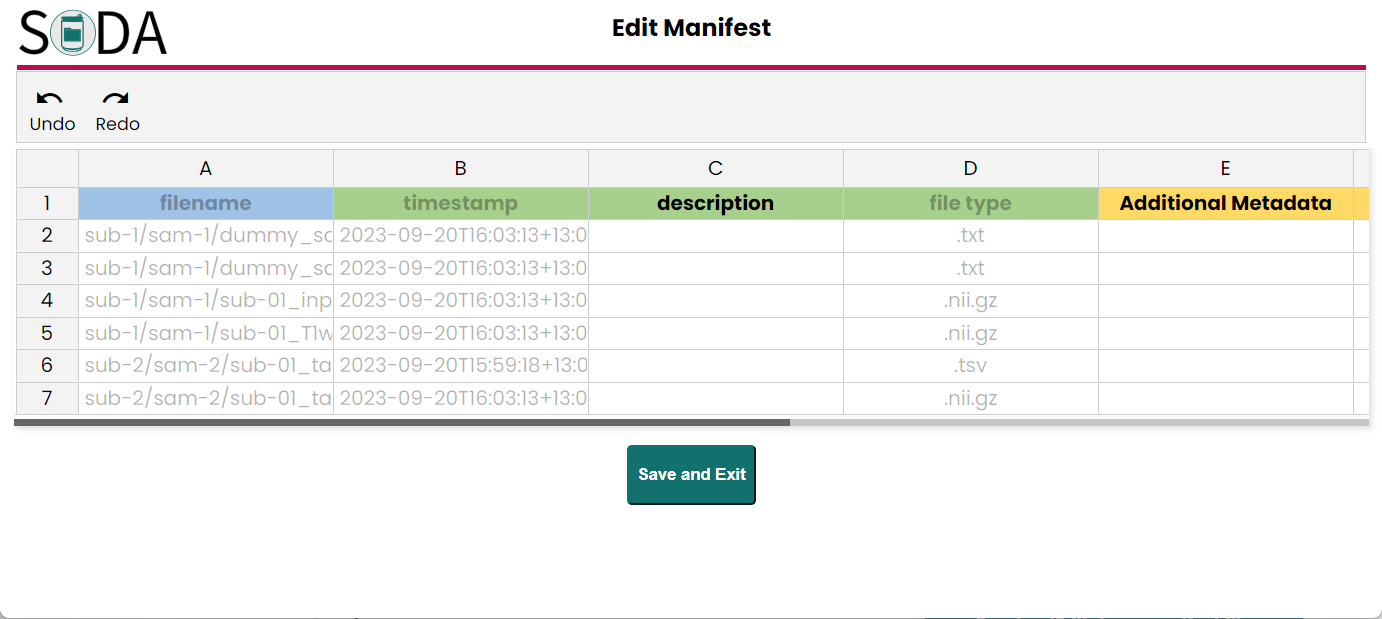
- Contributor information
- The SODA will ask user to fill the contributors information, these information will be automatically added into dataset_description metadata file. The definition for each role you can find in here.
- Also, in this step, each contributor must have a ORCID number, and one contributor role must be
PrincipalInvestigator. For others SODA also provide multiple roles for user to select.
Dataset Description
- You will be ask to fill in all elements in dataset description metadata except the contributor relevent elements.
- And all required element has a red star
*after the element, so you must fill that element. - As for the dataset keywords, user must give at least 3 keywords for the dataset.
Subjects and Sample metadata
- If you have similar subjects or samples in the dataset, you can quikly copy and paste the subjects and samples metadata information and edit them on SODA. This behavious is quite samilar to copy and paste the rows in the samples and subjects metadata files.
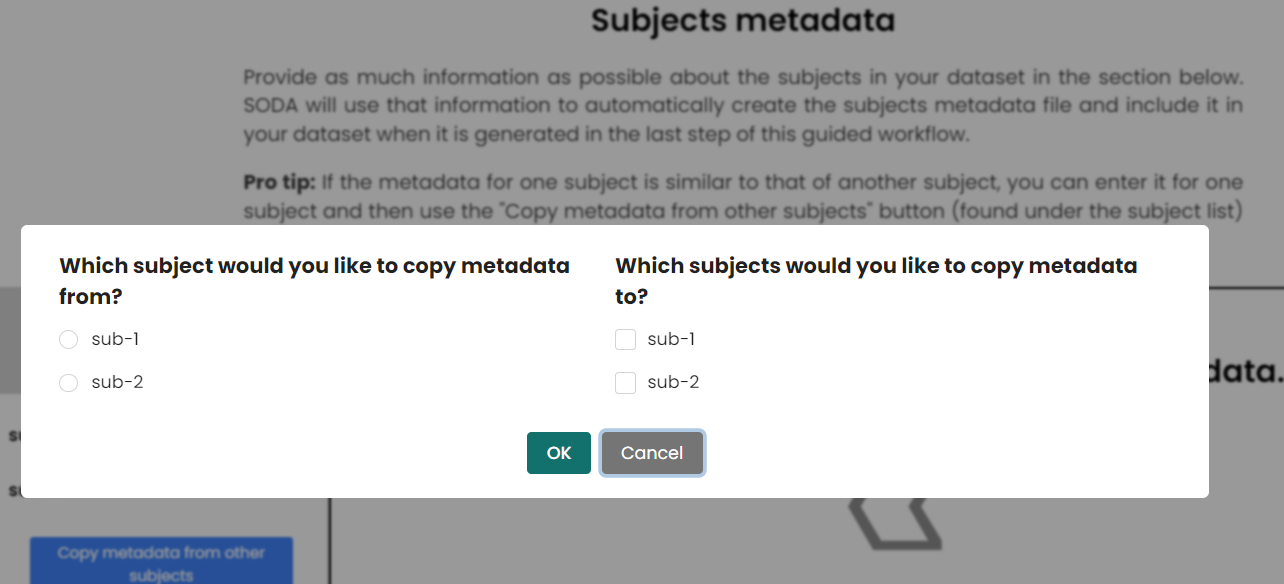
- If you have similar subjects or samples in the dataset, you can quikly copy and paste the subjects and samples metadata information and edit them on SODA. This behavious is quite samilar to copy and paste the rows in the samples and subjects metadata files.
Code metadata The
code_descriptionmetadata file is required for all datasets that include computation modeling related code files. It contains information that describes the code in terms of its quality. This file is typically prepared through osparc.io, an interactive, online simulation platform that hosts SPARC computational models and solvers.- User need to prepare the code_description metadata files on the osparc the upload into SODA.
- Also user can download the code_description metadata file and fill it out manually. Then upload to SODA.
README All SPARC datasets must include a README file.This is a plain text file that contains necessary details for the reuse of the data beyond that which is captured in the other metadata file. Some information that should be included is:
- How would a user use the files that are provided? e.g., first open file X and then look at file Y.
- What additional details do users need to know? Are some subjects missing data?
- Are there warnings about how to use the data or code?
- Are there appropriate/inappropriate uses for this data?
- Are there other places that users can go for more information? e.g., did you provide a GitHub repository or are there addional papers beyond what was provided in the metadata form?
Create SPARC SDS dataset via SODA Free Form Curation
NOTE: all empty folders will be deleted before generating the dataset since empty folders are not allowed in the SDS.
NOTE: If you are making only a few changes to an existing dataset (e.g., based on feedback from the SPARC Curation Team) Or you are very familar with the SPARC dataset structure and the metadata files and its elements, then you can choose to use the Free Form Curation to quickly generate a dataset.
NOTE: In the mode, if you want to create a new metadate file you can click the I want to start a new metadate_name.xlsx file, and if you want to edit current existing dataset (on you computer), you can choose I want to continue working on an existing metadate_name.xlsx file and upload it from your computer.
Create a new dataset
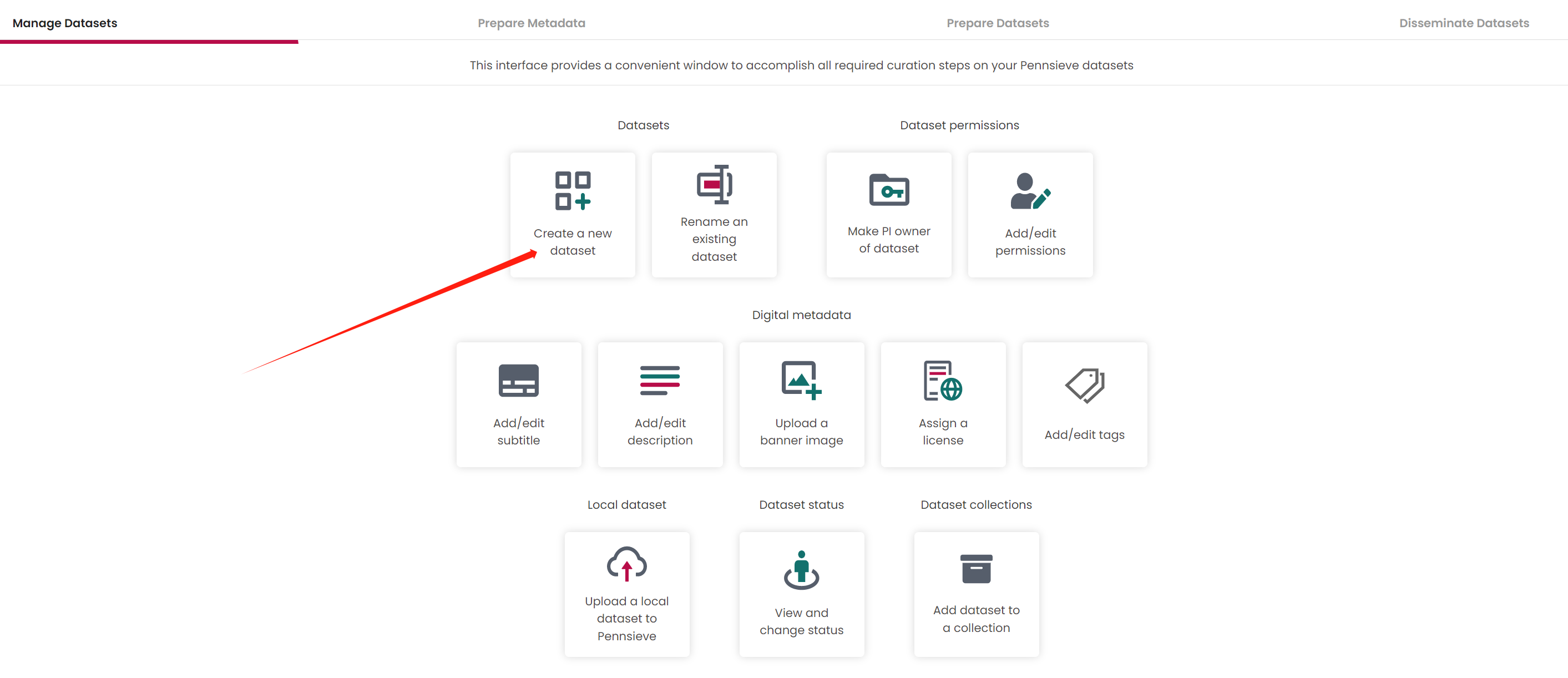
Fill out the dataset name
If there is an error appears and say Failed to change dataset status! Please select a vailda Pennsieve dataste . Just ignore it and Click the right-bottom Set PI owner of dataset button go next step.
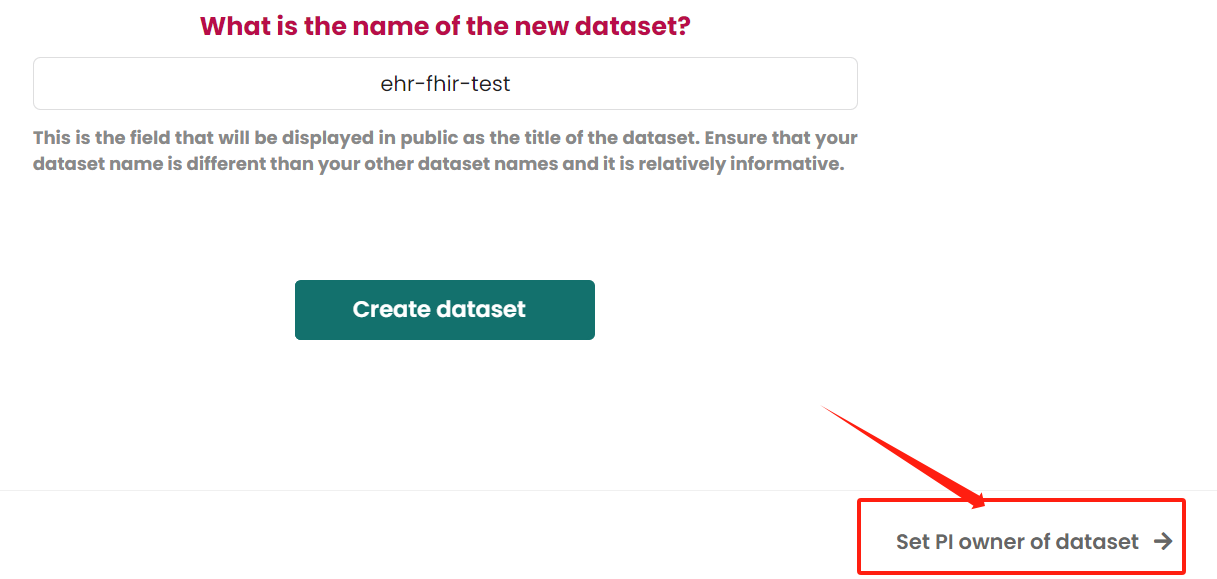
Make PI owner of dataset Guideline is here. If find it is not quite useful for your needs, then just click the next step at right-bottom
Set dataset permissionsbutton.Add permission for user
Guideline is here. If find it is not quite useful for your needs, then just click the next step at right-bottom Add a dataset subtitle button.
- Add dataset subtitle
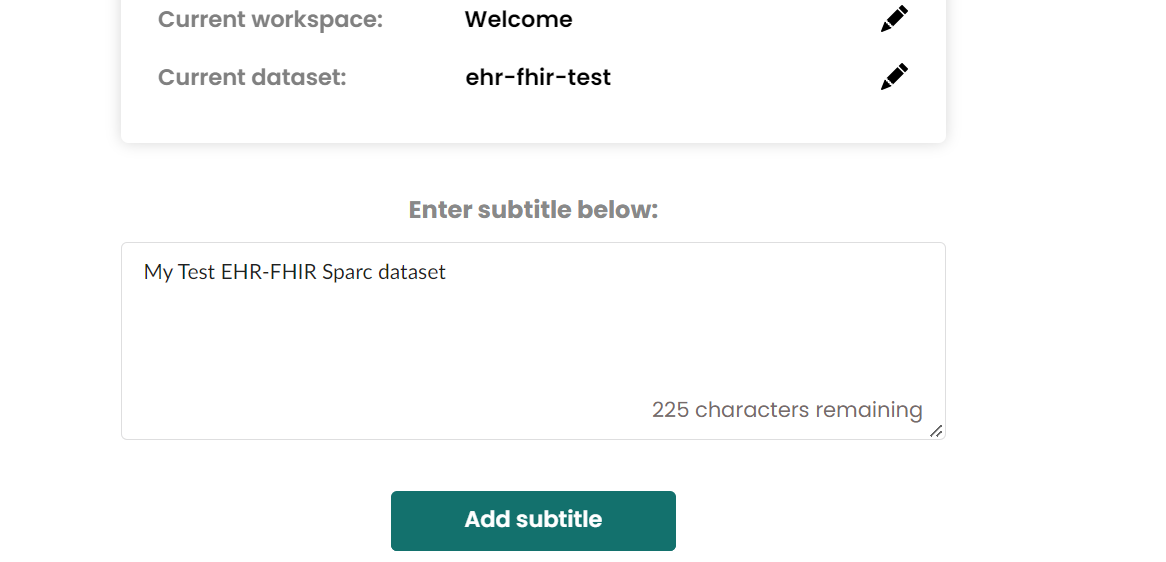
If have error with pennsieve relevant things, then just click the next step at right-bottom Add a dataset description button.
Add/edit description If have error with pennsieve relevant things, then just click the next step at right-bottom
Add a banner image to the datasetbutton.Upload a banner image If have error with pennsieve relevant things, then just click the next step at right-bottom
Assign a license to the datasetbutton.Add metadata files
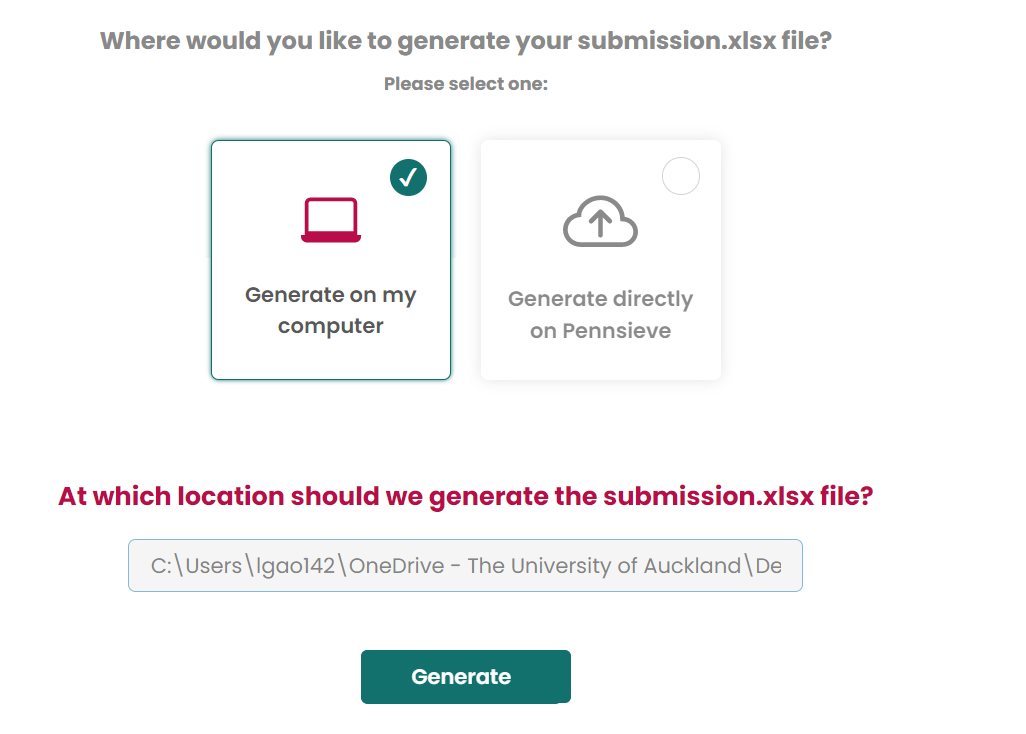
Create submission.xlsx
Choose
I want to start a new submission.xlsx file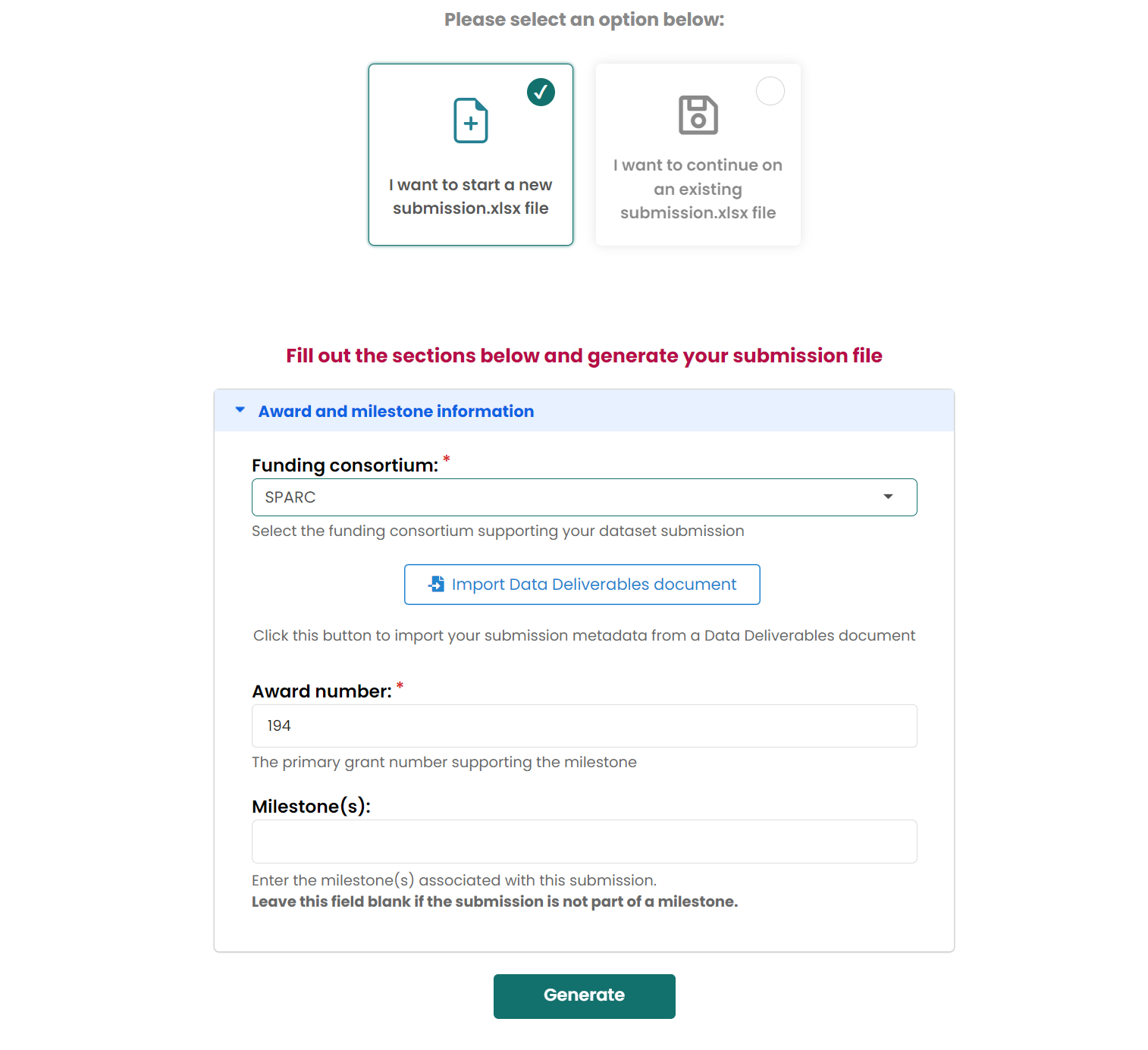
Then generate files on your computer.
Create dataset_description.xlsx, README.txt, subjects, samples, CHANGES.txt Same steps as above!
Organize dataset Choose
Prepare a new dataset, if you are trying to generate a new dataset.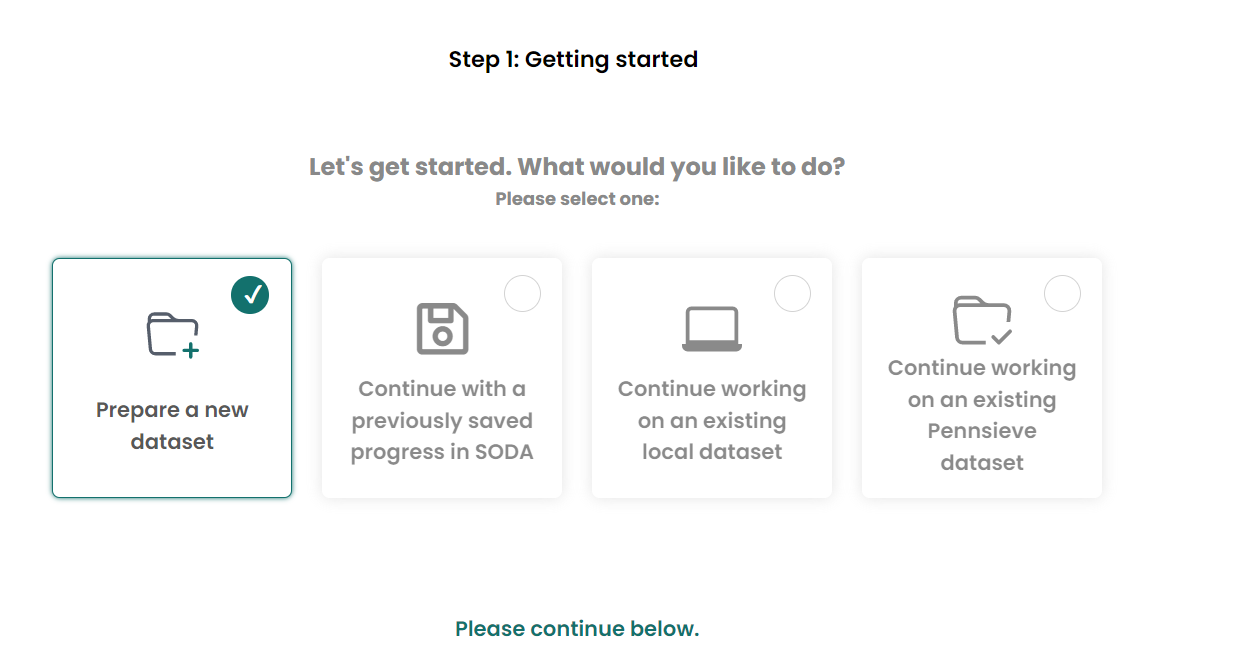
- Choose which category folders you would like to have them in the dataset NOTE: after you choose the folders, you must upload files into that folder, otherwise the SODA won't generate that folder for your dataset.

Upload files into the category folder that you select above Double click the folder in SODA, then upload files for that folder.
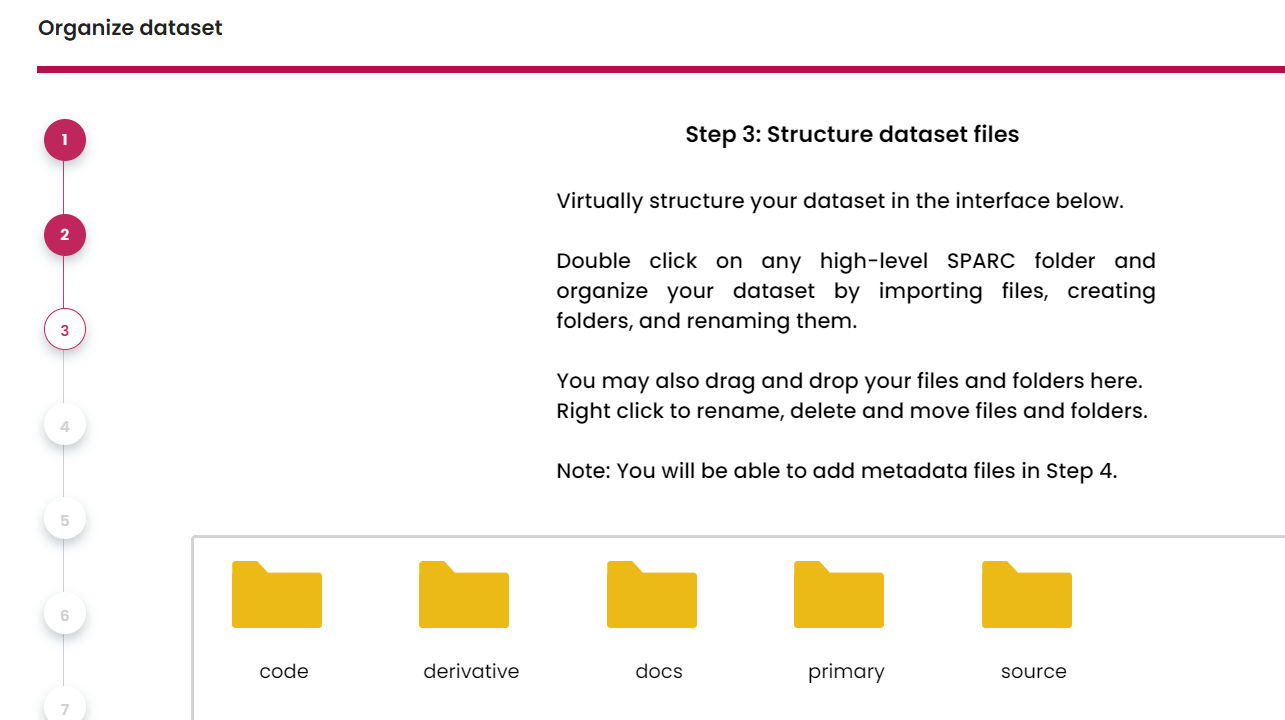
Upload the metadata files (You generated them at
Add metadata files step), click the file name to upload,

- Allow SODA to auto-generate manifest files
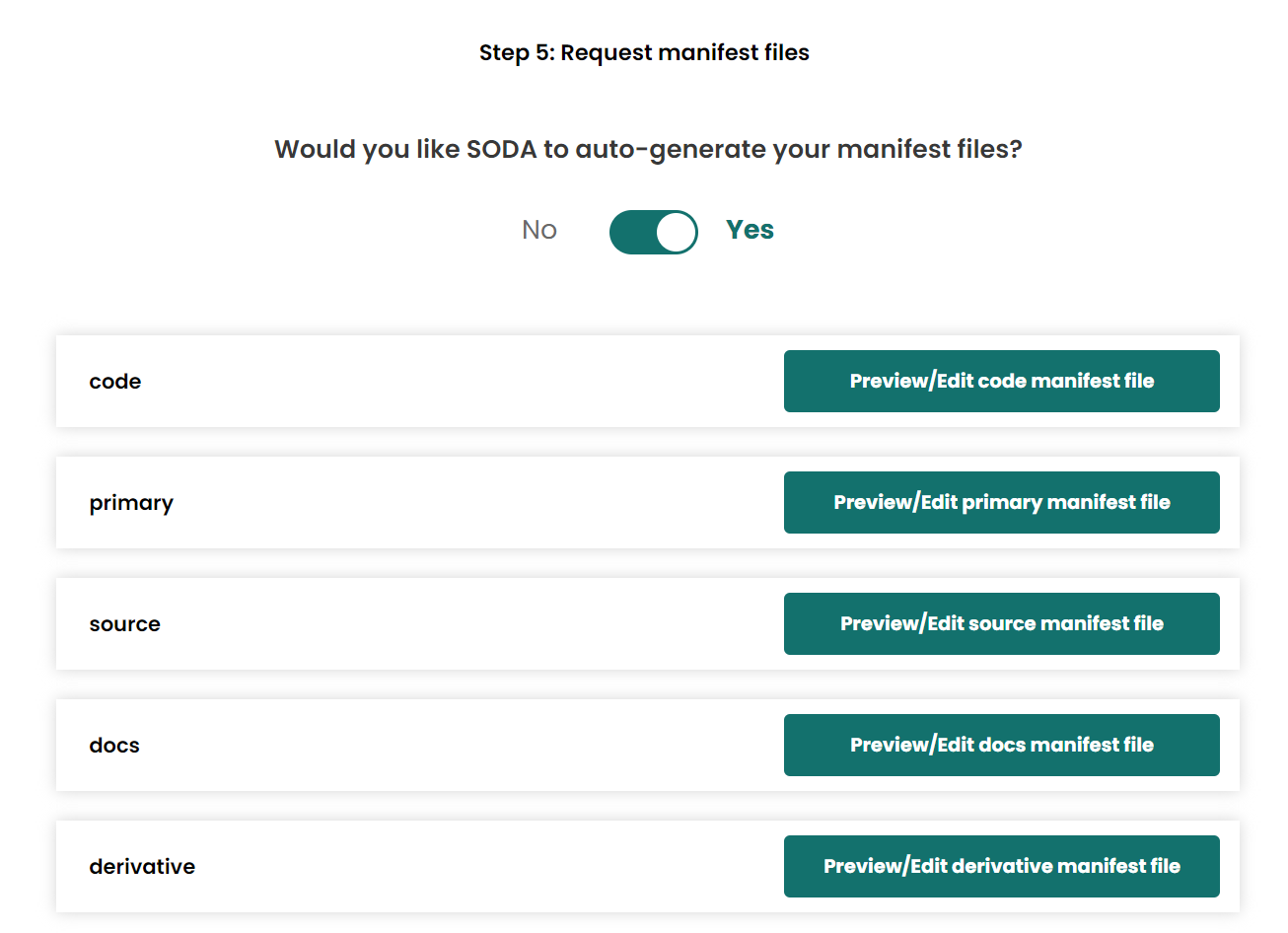
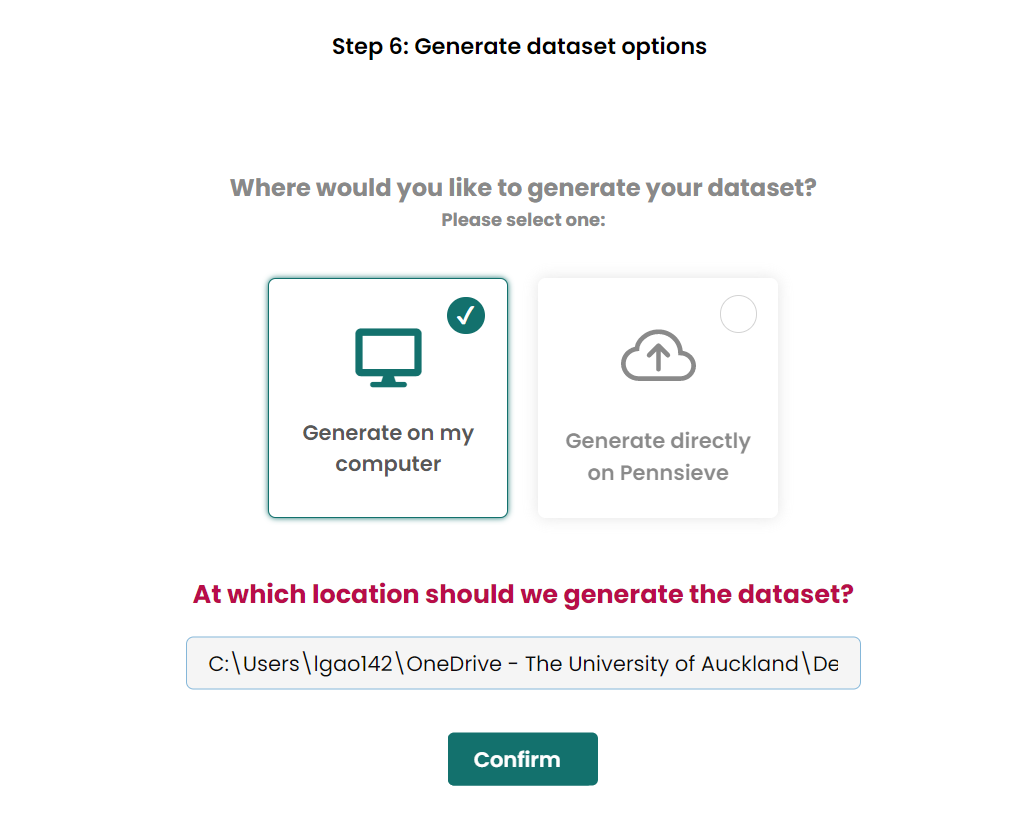
- Generate dataset on compute, the same loaction as the
Add metadata filestep.
- Skip the validation step.
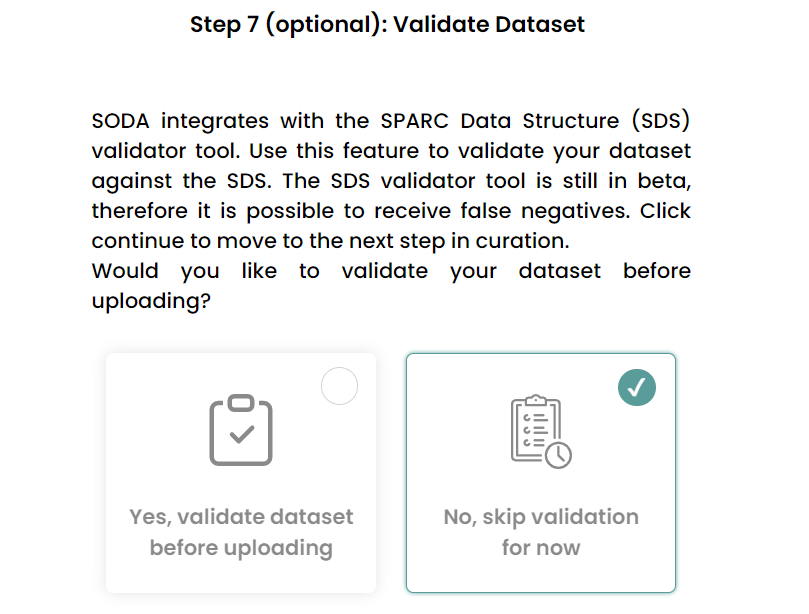
Generate the dataset.
Check the dataset on your computer, then manunaly copy and paste the category files into each category folder in dataset. Because the SODA won't generate the files for you, they just use the file names to generate the manifest metadata files for your dataset.
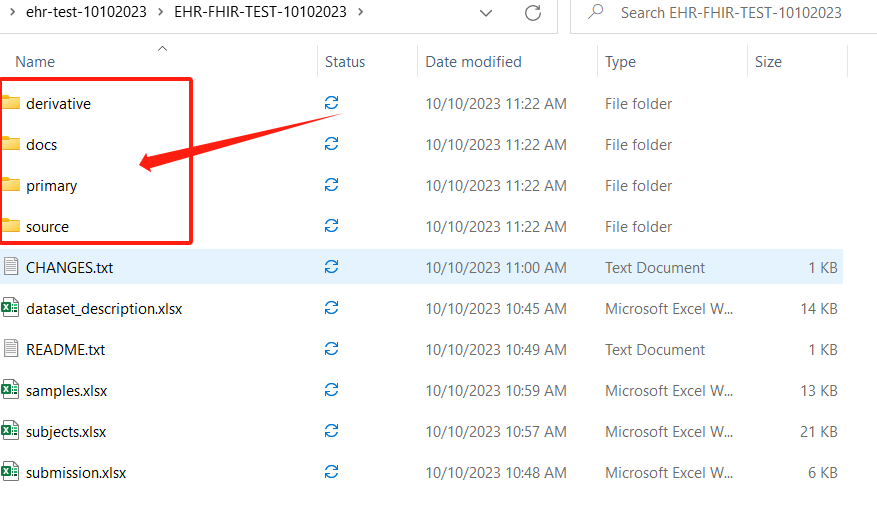
The difference between Sparc-me and SODA dataset
Dataset Version
- Sparc-me, v2.0.0
- SODA, v2.1.0
Manifest metadata file
- Sparc-me, only have one manifest file in the dataset root folder, it contains all categories(
primary,derivative,docs,code,protocolandsource) infomations. - SODA, generate manifest files for each category, and store the manifest file into each category folder.
Dataset description
- contributor roles:
- Sparc-me hasn't specific the role names. Allow user to manually typing the role name.
- SODA provide multiple role names for user to choose, and it has rules:
- User must choose roles from they provided.
- One dataset must contain a
PrincipalInvestigatorrole.
- ORCID number
- Sparc-me hasn't check if the number is a valid ORCID number.
- SODA will validate the number, if not correct won't allow go next steps.
- Keywords
- Sparc-me at least one and above.
- SODA at least three and above.
Subjects and Samples
- They are quite same.
- Sparc-me provide the Sample and Subject class to represent as each row in the subjects and samples metdata. Allow user quickly to edit them based on Sample and Subject class programme functions.
- SODA, allow user to copy a similar Sample or Subject infomations to another one. It is also edit each row information in the samples and subjects metadata files.
- Subject
pool idandsubject experimental group:- Sparc-me: no related functions or guidelines for them.
- SOAD: they are related. And it has a lot of guidelines/examples for each elements to remind users to fill out.
Thumbnails
- In Sparc-me its called thumbnails and stored in the
docsfolder. Haven't checked the image format. - In SODA its called banner, only
.PNG, .JPEG, .TIFFcan be used.
Pool, Subject and Sample
- Both Sparc-me and SODA start the subject and sample names with
sub-andsam-, but they have difference. - Sparc-me, it don't have pool related folders, and the subject and sample names are only unique under their parent folder, if they are under different parent folder their names are no longer unique. E.g,
sam-1undersub-1andsam-1undersub-2 - SODA, it has pool layer, the pool folder is the subject parent folder. And every names in SODA are unique. E.g, if
sub-1underpool-1andsam-1undersub-1, then thesub-1cannot underpool-xandsam-1cannot undersub-x.
Empty category folders (primary , derivative, docs, code, protocol and source)
- Sparc-me. allow them exist
- SODA, will delete them when generate a new dataset. Because empty folders are not allowed in SDS.